Abstract
Ryanodine receptors (RyRs) mediate the rapid release of calcium (Ca2+) from intracellular stores into the cytosol, which is essential for numerous cellular functions including excitation–contraction coupling in muscle. Lack of sufficient structural detail has impeded understanding of RyR gating and regulation. Here we report the closed-state structure of the 2.3-megadalton complex of the rabbit skeletal muscle type 1 RyR (RyR1), solved by single-particle electron cryomicroscopy at an overall resolution of 4.8 Å. We fitted a polyalanine-level model to all 3,757 ordered residues in each protomer, defining the transmembrane pore in unprecedented detail and placing all cytosolic domains as tertiary folds. The cytosolic assembly is built on an extended α-solenoid scaffold connecting key regulatory domains to the pore. The RyR1 pore architecture places it in the six-transmembrane ion channel superfamily. A unique domain inserted between the second and third transmembrane helices interacts intimately with paired EF-hands originating from the α-solenoid scaffold, suggesting a mechanism for channel gating by Ca2+.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
Cryo-EM reconstructions of RyR1 have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-6106 (EGTA-treated RyR1) and EMD-6107 (EGTA- and CIP-treated RyR1). The Cα coordinates of the model have been deposited in the Protein Data Bank (PDB) under accession number 3J8E.
References
Zalk, R., Lehnart, S. E. & Marks, A. R. Modulation of the ryanodine receptor and intracellular calcium. Annu. Rev. Biochem. 76, 367–385 (2007)
Marx, S. O. et al. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell 101, 365–376 (2000)
Paolini, C., Protasi, F. & Franzini-Armstrong, C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. J. Mol. Biol. 342, 145–153 (2004)
Meissner, G. & Lu, X. Dihydropyridine receptor-ryanodine receptor interactions in skeletal muscle excitation-contraction coupling. Biosci. Rep. 15, 399–408 (1995)
Stern, M. D., Pizarro, G. & Rios, E. Local control model of excitation-contraction coupling in skeletal muscle. J. Gen. Physiol. 110, 415–440 (1997)
Marx, S. O., Ondrias, K. & Marks, A. R. Coupled gating between individual skeletal muscle Ca2+ release channels (ryanodine receptors). Science 281, 818–821 (1998)
Wehrens, X. H. et al. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell 113, 829–840 (2003)
Liu, X. et al. Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell 150, 1055–1067 (2012)
Andersson, D. C. et al. Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metab. 14, 196–207 (2011)
Bellinger, A. M. et al. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nature Med. 15, 325–330 (2009)
Marks, A. R. Calcium cycling proteins and heart failure: mechanisms and therapeutics. J. Clin. Invest. 123, 46–52 (2013)
Andersson, D. C. & Marks, A. R. Fixing ryanodine receptor Ca leak — a novel therapeutic strategy for contractile failure in heart and skeletal muscle. Drug Discov. Today Dis. Mech. 7, e151–e157 (2010)
Serysheva, I. I. et al. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl Acad. Sci. USA 105, 9610–9615 (2008)
Fernández, I. S. et al. Molecular architecture of a eukaryotic translational initiation complex. Science 342, 1240585 (2013)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Groves, M. R. & Barford, D. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9, 383–389 (1999)
Xiong, L., Zhang, J. Z., He, R. & Hamilton, S. L. A. Ca2+-binding domain in RyR1 that interacts with the calmodulin binding site and modulates channel activity. Biophys. J. 90, 173–182 (2006)
Tung, C. C., Lobo, P. A., Kimlicka, L. & Van Petegem, F. The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature 468, 585–588 (2010)
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010)
Groves, M. R. & Barford, D. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9, 383–389 (1999)
Huber, A. H., Nelson, W. J. & Weis, W. I. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90, 871–882 (1997)
Yu, F. H., Yarov-Yarovoy, V., Gutman, G. A. & Catterall, W. A. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol. Rev. 57, 387–395 (2005)
Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W. A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011)
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013)
Owsianik, G., Talavera, K., Voets, T. & Nilius, B. Permeation and selectivity of TRP channels. Annu. Rev. Physiol. 68, 685–717 (2006)
Tripathy, A., Xu, L., Mann, G. & Meissner, G. Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor). Biophys. J. 69, 106–119 (1995)
Volkov, A. G., Paula, S. & Deamer, D. W. Two mechanisms of permeation of small neutral molecules and hydrated ions across phospholipid bilayers. Bioelectrochem. Bioenerg. 42, 153–160 (1997)
Jiang, Y. et al. X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 (2003)
Bezprozvanny, I., Watras, J. & Ehrlich, B. E. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature 351, 751–754 (1991)
Ludtke, S. J., Serysheva, I. I., Hamilton, S. L. & Chiu, W. The pore structure of the closed RyR1 channel. Structure 13, 1203–1211 (2005)
Samsó, M., Wagenknecht, T. & Allen, P. D. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nature Struct. Mol. Biol. 12, 539–544 (2005)
Yuchi, Z., Lau, K. & Van Petegem, F. Disease mutations in the ryanodine receptor central region: crystal structures of a phosphorylation hot spot domain. Structure 20, 1201–1211 (2012)
Sharma, P. et al. Structural determination of the phosphorylation domain of the ryanodine receptor. FEBS J. 279, 3952–3964 (2012)
Marks, A. R., Fleischer, S. & Tempst, P. Surface topography analysis of the ryanodine receptor/junctional channel complex based on proteolysis sensitivity mapping. J. Biol. Chem. 265, 13143–13149 (1990)
Yin, C. C., Han, H., Wei, R. & Lai, F. A. Two-dimensional crystallization of the ryanodine receptor Ca2+ release channel on lipid membranes. J. Struct. Biol. 149, 219–224 (2005)
Woo, J.-S. et al. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 25, 1353–1363 (2006)
Brillantes, A. B. et al. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell 77, 513–523 (1994)
Gaburjakova, M. et al. FKBP12 binding modulates ryanodine receptor channel gating. J. Biol. Chem. 276, 16931–16935 (2001)
Lau, K. & van Petegem, F. Crystal structures of wild type and disease mutant forms of the ryanodine receptor SPRY2 domain. Nature Comm. 5, 5937 (2014)
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004)
Xin, H. B., Timerman, A. P., Onoue, H., Wiederrecht, G. J. & Fleischer, S. Affinity purification of the ryanodine receptor/calcium release channel from fast twitch skeletal muscle based on its tight association with FKBP12. Biochem. Biophys. Res. Commun. 214, 263–270 (1995)
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988)
Wagenknecht, T., Frank, J., Boublik, M., Nurse, K. & Ofengand, J. Direct localization of the tRNA–anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. J. Mol. Biol. 203, 753–760 (1988)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 http://dx.doi.org/10.1006/jsbi.1996.0030 (1996)
Langlois, R. et al. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. J. Struct. Biol. 186, 1–7 (2014)
Scheres, S. H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012)
Reiken, S. et al. PKA phosphorylation activates the calcium release channel (ryanodine receptor) in skeletal muscle: defective regulation in heart failure. J. Cell Biol. 160, 919–928 (2003)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003)
Pintilie, G. D., Zhang, J., Goddard, T. D., Chiu, W. & Gossard, D. C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
McGuffin, L. J., Bryson, K. & Jones, D. T. The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 (2000)
Acknowledgements
We thank R. Axel for discussions and M. Thomas for assistance with the preparation of illustrations. This work was supported by grants from the National Institutes of Health (R01AR060037 and R01HL061503 to A.R.M., U54GM095315 to W.A.H., R01GM29169 to J.F.), and the Howard Hughes Medical Institute (to J.F.). R.Z. was a fellow of the American Heart Association (0625919T), and O.B.C. was supported by an overseas biomedical fellowship (NHMRC; Australia) and a Charles H. Revson Senior Fellowship (Charles H. Revson Foundation).
Author information
Authors and Affiliations
Contributions
A.R.M. conceived the project, designed experiments, analysed data and wrote the manuscript. W.A.H. conceived the project, designed experiments, analysed data and wrote the manuscript. J.F. designed the cryo-EM experiments, analysed data and wrote the manuscript. R.Z. designed experiments, isolated the RyR1 used in this work, analysed data and wrote the manuscript. O.B.C. designed experiments, analysed data, built the atomic model and wrote the manuscript. F.M. designed experiments. A.G. designed experiments and conducted the cryo-EM experiments, including data acquisition and processing, and wrote the manuscript. R.A.G. conducted the cryo-EM experiments. S.R. conducted phospho-RyR immunoblot experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Skeletal muscle RyR1 purification.
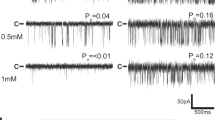
a, Coomassie blue staining of SDS–PAGE showing molecular weight standards (MWS), CHAPS-solubilized sarcoplasmic reticulum membrane (SR), RyR1 eluted with calstabin2 from a glutathione S-transferase (GST)–calstabin1 affinity chromatography column (AC), the eluted RyR1 from fast protein liquid chromatography (FPLC) size-exclusion chromatograph (SEC). b, FPLC plot showing the RyR1 peak at ∼7 ml elution and the excess calstabin2 (Cs2) peak at ∼12 ml elution. c, Immunoblot analysis of CIP-treated RyR1 probed at indicated time points with (from bottom) anti-RyR1 (34C) antibody, anti-phosphotyrosine antibody (pTyr; Abcam ab10321), anti-phosphothreonine antibody (pThr; Abcam ab79851), anti-phosphoserine antibody (pSer; Abcam ab9332) and anti-RyR phospho-specific antibody (RyR1-pS2843) that recognizes the PKA phosphorylated site on RyR1.
Extended Data Figure 2 Particle picking and two-dimensional class averages.
a, Sample micrograph of the RyR1-CIP-EGTA data set after motion correction, with red boxes around the particles picked by Autopicker47. Scale bar, 500 Å. b, Sample power spectrum of a twice-decimated micrograph after motion correction. c, Euler angle distribution before symmetry was imposed of the particles that went into the CIP-treated data set final reconstruction. Latitude (radial distance) corresponds to θ from 0 to 90°. Longitude (position on the circle) corresponds to ϕ, from 0 to 360°. The dots colour and area represent the number of particles in each view. d, Two-dimensional projections of the final CIP-treated map (upper rows; red lines) compared to their respective reference-free two-dimensional class averages (lower rows).
Extended Data Figure 3 Classification and protomer boundaries.
a, Classification of the RyR1-EGTA data set. First row, refined volume with all particles. Second row, primary classification with a number of classes (K = 10) giving rise to two major classes, one refined to 4.8 Å (blue box) and one refined to 5.0 Å (green box). This class (green box) was subclassified with K = 10 (third row), and yielded one class with a missing or disordered cytosolic portion of a protomer (red box). b, Classification of the RyR1-EGTA-CIP-treated data set with K = 8. Class 8 (purple box) was refined to 5.0 Å. c, Views from the cytosol, membrane plane and lumen of the RyR1 model superimposed with a difference map between the full tetramer map (blue box) and the map with the cytosolic region of one protomer missing or disordered (red box).
Extended Data Figure 4 RyR1 cryo-EM local resolution map.
a, Gold-standard FSC curve for the three-dimensional reconstructions, marked with resolutions corresponding to FSC = 0.143. b, Cytosolic, membrane plane and luminal views of RyR1 (EGTA- and CIP-treated data set) local resolution distribution from 4 (blue) to 6 (red) Å resolution. c, Local resolution distribution through a slab of density coincident with channel axis. d, Same slab as c for the EGTA-treated without CIP. e, Slices through the volume of the CIP-EGTA data set (top) and EGTA data set (bottom). Slice direction and number are indicated on the images.
Extended Data Figure 5 Representative densities of RyR1-selected regions.
a–e, Representative density (grey mesh) in selected regions of the map. The protomers are represented as Cα traces, in different colours for clarity, with enlarged views of the following regions: calstabin2 (a), the bridge solenoid (b), NTD (c), the pore region (d) and S6 (e).
Extended Data Figure 6 RyR1 local model to map correlation.
a, Cytosolic, membrane plane and luminal views of the local correlation (calculated in a 5 × 5 voxel sliding window) between a map calculated from the model (filtered to 5 Å) and the density map of dephosphorylated RyR1, depicted in spectral colouring from 0.7 (red) to 1 (blue). b, c, Local model/density map correlation within a slab of density through the plane of the membrane, highlighting the unmodelled rod of density on the periphery of the transmembrane region (b) and a slab coinciding with the channel axis (c).
Extended Data Figure 7 α- solenoid subdomains.
RyR1 density map (grey semitransparent surface) superimposed with the α-solenoid scaffold of RyR1. a, Core of the α-solenoid scaffold (insertions and elaborations not shown). Green, bridging solenoid; blue, NTD solenoid; red, core solenoid. b, Alignment of NTD with an α-solenoid structure (PDB code 3NMX). c, Alignment of core solenoid with an α-solenoid structure (PDB code 1G3J). d, Overlay of bridging solenoid with an α-solenoid structure (PDB code 1WA5). In b–d, RyR1 α-solenoid repeats are depicted in spectral colouring from blue (N terminus) to red (C terminus), and the aligned α-solenoid protein is represented in dark grey.
Extended Data Figure 8 Architecture of bridging and core solenoids.
a, Density map of RyR1 in dark blue mesh superimposed with the bridging solenoid shown in detail on right, as labelled. b, Two views of the interaction of the core solenoid (spectral colouring) containing the putative Ca2+-binding domain with CTD (grey) as labelled.
Extended Data Figure 9 Calstabin2-binding site.
Views in the membrane plane and cytosol of RyR1 with enlarged views of calstabin2 (yellow) bound to RyR1. SPRY1 is depicted in light blue, SPRY2 in cyan, the bridging solenoid in green and the calstabin binding helix in purple.
Extended Data Figure 10 Putative Ca2+-binding domain in RyR1.
a, Sequence alignment of rabbit RyR1–3 with the C-lobe of human calmodulin (hCaM). b, Structural alignment of the C-lobe of yeast calmodulin with the model of RyR1.
Rights and permissions
About this article
Cite this article
Zalk, R., Clarke, O., des Georges, A. et al. Structure of a mammalian ryanodine receptor. Nature 517, 44–49 (2015). https://doi.org/10.1038/nature13950
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13950
This article is cited by
-
Identification, characterization and spatiotemporal expression analysis of the FKBP family genes in Locusta migratoria
Scientific Reports (2023)
-
Heart failure-induced cognitive dysfunction is mediated by intracellular Ca2+ leak through ryanodine receptor type 2
Nature Neuroscience (2023)
-
THADA inhibition in mice protects against type 2 diabetes mellitus by improving pancreatic β-cell function and preserving β-cell mass
Nature Communications (2023)
-
The modes of action of ion-channel-targeting neurotoxic insecticides: lessons from structural biology
Nature Structural & Molecular Biology (2023)
-
Biophysical reviews top five: voltage-dependent charge movement in nerve and muscle
Biophysical Reviews (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.