Abstract
Bestrophin calcium-activated chloride channels (CaCCs) regulate the flow of chloride and other monovalent anions across cellular membranes in response to intracellular calcium (Ca2+) levels. Mutations in bestrophin 1 (BEST1) cause certain eye diseases. Here we present X-ray structures of chicken BEST1–Fab complexes, at 2.85 Å resolution, with permeant anions and Ca2+. Representing, to our knowledge, the first structure of a CaCC, the eukaryotic BEST1 channel, which recapitulates CaCC function in liposomes, is formed from a pentameric assembly of subunits. Ca2+ binds to the channel’s large cytosolic region. A single ion pore, approximately 95 Å in length, is located along the central axis and contains at least 15 binding sites for anions. A hydrophobic neck within the pore probably forms the gate. Phenylalanine residues within it may coordinate permeating anions via anion–π interactions. Conformational changes observed near the ‘Ca2+ clasp’ hint at the mechanism of Ca2+-dependent gating. Disease-causing mutations are prevalent within the gating apparatus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hartzell, C., Putzier, I. & Arreola, J. Calcium-activated chloride channels. Annu. Rev. Physiol. 67, 719–758 (2005)
Caputo, A. et al. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322, 590–594 (2008)
Schroeder, B. C., Cheng, T., Jan, Y. N. & Jan, L. Y. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134, 1019–1029 (2008)
Yang, Y. et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455, 1210–1215 (2008)
Sun, H., Tsunenari, T., Yau, K.-W. & Nathans, J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc. Natl Acad. Sci. USA 99, 4008–4013 (2002)
Tsunenari, T., Nathans, J. & Yau, K.-W. Ca2+-activated Cl− current from human bestrophin-4 in excised membrane patches. J. Gen. Physiol. 127, 749–754 (2006)
Hartzell, H. C., Qu, Z., Yu, K., Xiao, Q. & Chien, L.-T. Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiol. Rev. 88, 639–672 (2008)
Kunzelmann, K. et al. Role of the Ca2+ -activated Cl− channels bestrophin and anoctamin in epithelial cells. Biol. Chem. 392, 125–134 (2011)
Gomez, N. M., Tamm, E. R. & Straubeta, O. Role of bestrophin-1 in store-operated calcium entry in retinal pigment epithelium. Pflugers Archiv. 465, 481–495 (2013)
Tsunenari, T. et al. Structure-function analysis of the bestrophin family of anion channels. J. Biol. Chem. 278, 41114–41125 (2003)
Qu, Z., Fischmeister, R. & Hartzell, C. Mouse bestrophin-2 is a bona fide Cl− channel: identification of a residue important in anion binding and conduction. J. Gen. Physiol. 123, 327–340 (2004)
Chien, L.-T., Zhang, Z.-R. & Hartzell, H. C. Single Cl− channels activated by Ca2+ in Drosophila S2 cells are mediated by bestrophins. J. Gen. Physiol. 128, 247–259 (2006)
Xiao, Q., Prussia, A., Yu, K., Cui, Y.-y. & Hartzell, H. C. Regulation of bestrophin Cl channels by calcium: role of the C terminus. J. Gen. Physiol. 132, 681–692 (2008)
Marquardt, A. et al. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease). Hum. Mol. Genet. 7, 1517–1525 (1998)
Petrukhin, K. et al. Identification of the gene responsible for Best macular dystrophy. Nature Genet. 19, 241–247 (1998)
Davidson, A. E. et al. Missense mutations in a retinal pigment epithelium protein, Bestrophin-1, cause retinitis pigmentosa. Am. J. Hum. Genet. 85, 581–592 (2009)
Boon, C. J. et al. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog. Retin. Eye Res. 28, 187–205 (2009)
Xiao, Q., Hartzell, H. C. & Yu, K. Bestrophins and retinopathies. Pflügers Archiv. 460, 559–569 (2010)
Kinnick, T. R. et al. Autosomal recessive vitelliform macular dystrophy in a large cohort of vitelliform macular dystrophy patients. Retina 31, 581–595 (2011)
Wittström, E., Ponjavic, V., Bondeson, M. L. & Andreasson, S. Anterior segment abnormalities and angle-closure glaucoma in a family with a mutation in the BEST1 gene and Best vitelliform macular dystrophy. Ophthalmic Genet. 32, 217–227 (2011)
Yu, K., Cui, Y. & Hartzell, H. C. The bestrophin mutation A243V, linked to adult-onset vitelliform macular dystrophy, impairs its chloride channel function. Invest. Ophthalmol. Vis. Sci. 47, 4956–4961 (2006)
Yu, K., Qu, Z., Cui, Y. & Hartzell, H. C. Chloride channel activity of bestrophin mutants associated with mild or late-onset macular degeneration. Invest. Ophthal. Vis. Sci. 48, 4694–4705 (2007)
Marchant, D. et al. New VMD2 gene mutations identified in patients affected by Best vitelliform macular dystrophy. J. Med. Genet. 44, e70 (2007)
Milenkovic, V. M., Röhrl, E., Weber, B. H. F. & Strauss, O. Disease-associated missense mutations in bestrophin-1 affect cellular trafficking and anion conductance. J. Cell Sci. 124, 2988–2996 (2011)
Bharill, S., Fu, Z., Palty, R. & Isacoff, E. Y. Stoichiometry and specific assembly of Best ion channels. Proc. Natl Acad. Sci. USA 111, 6491–6496 (2014)
Qu, Z. Q., Yu, K., Cui, Y.-y., Ying, C. & Hartzell, C. Activation of bestrophin Cl− channels is regulated by C-terminal domains. J. Biol. Chem. 282, 17460–17467 (2007)
Kranjc, A. et al. Regulation of bestrophins by Ca2+: a theoretical and experimental study. PLoS ONE 4, e4672 (2009)
Qu, Z. & Hartzell, H. C. Bestrophin Cl− channels are highly permeable to HCO3−. Am. J. Physiol. Cell Physiol. 294, C1371–C1377 (2008)
Stotz, S. C. & Clapham, D. E. Anion-sensitive fluorophore identifies the Drosophila swell-activated chloride channel in a genome-wide RNA interference screen. PLoS ONE 7, e46865 (2012)
Lee, S. et al. Channel-mediated tonic GABA release from glia. Science 330, 790–796 (2010)
Woo, D. H. et al. TREK-1 and Best1 channels mediate fast and slow glutamate release in astrocytes upon GPCR activation. Cell 151, 25–40 (2012)
O’Driscoll, K. E., Leblanc, N., Hatton, W. J. & Britton, F. C. Functional properties of murine bestrophin 1 channel. Biochem. Biophys. Res. Commun. 384, 476–481 (2009)
Qu, Z. & Hartzell, C. Determinants of anion permeation in the second transmembrane domain of the mouse bestrophin-2 chloride channel. J. Gen. Physiol. 124, 371–382 (2004)
Qu, Z., Chien, L.-T., Cui, Y. & Hartzell, H. C. The anion-selective pore of the bestrophins, a family of chloride channels associated with retinal degeneration. J. Neurosci. 26, 5411–5419 (2006)
Gifford, J. L., Walsh, M. P. & Vogel, H. J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 405, 199–221 (2007)
Yuan, P., Leonetti, M. D., Hsiung, Y. & MacKinnon, R. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature 481, 94–97 (2012)
Dutzler, R., Campbell, E. B., Cadene, M., Chait, B. T. & MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002)
Hibbs, R. E. & Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011)
Hille, B. Ionic Channels of Excitable Membranes 2nd edn (Sinauer Associates, 1992)
Dougherty, D. A. Cation-π interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271, 163–168 (1996)
Jackson, M. R. et al. A preference for edgewise interactions between aromatic rings and carboxylate anions: the biological relevance of anion-quadrupole interactions. J. Phys. Chem. B 111, 8242–8249 (2007)
Philip, V. et al. A survey of aspartate-phenylalanine and glutamate-phenylalanine interactions in the protein data bank: searching for anion-π pairs. Biochemistry 50, 2939–2950 (2011)
Thomas, K. A., Smith, G. M., Thomas, T. B. & Feldmann, R. J. Electronic distributions within protein phenylalanine aromatic rings are reflected by the three-dimensional oxygen atom environments. Proc. Natl Acad. Sci. USA 79, 4843–4847 (1982)
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006)
Kilmartin, J. V., Wright, B. & Milstein, C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93, 576–582 (1982)
Long, S. B., Campbell, E. B. & MacKinnon, R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 (2005)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Cowtan, K. D. ‘dm': An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Prot. Crystallogr. 31, 34–38 (1994)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Brunger, A. T. Version 1.2 of the crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Konecny, R., Baker, N. A. & McCammon, J. A. iAPBS: a programming interface to Adaptive Poisson-Boltzmann Solver (APBS). Comp. Sci. Discov. 5, 015005 (2012)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360,–376 (1996)
Miller, A. N. & Long, S. B. Crystal structure of the human two-pore domain potassium channel K2P1. Science 335, 432–436 (2012)
Hou, X., Pedi, L., Diver, M. M. & Long, S. B. Crystal structure of the calcium release-activated calcium channel Orai. Science 338, 1308–1313 (2012)
Lee, S.-Y., Letts, J. A. & MacKinnon, R. Functional reconstitution of purified human Hv1 H+ channels. J. Mol. Biol. 387, 1055–1060 (2009)
Tsien, R. & Pozzan, T. Measurement of cytosolic free Ca2+ with quin2. Methods Enzymol. 172, 230–262 (1989)
Schoenmakers, T. J., Visser, G. J., Flik, G. & Theuvenet, A. P. CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. Biotechniques 12, 870–874,–876–879 (1992)
Acknowledgements
We acknowledge the staff at beamlines X25 and X29 of the National Synchrotron Light Source, beamline 24-ID-C of the Advanced Photon Source, and F. Weis-Garcia and the Monoclonal Antibody Facility at MSKCC. We thank C. Lima, M. Long, N. Pavletich, V. Ruta and members of the laboratory for discussions. S.B.L. is a recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences.
Author information
Authors and Affiliations
Contributions
All authors contributed to project design and performed experiments. V.K.D. and S.B.L. determined structures. S.B.L. wrote the manuscript with contributions from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
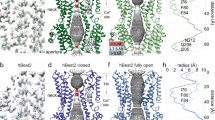
Extended Data Figure 1 Sequence alignment and secondary structure.
The amino acid sequences of the crystallized chicken (Gallus gallus) BEST1 construct (amino acids 2–405) and human BEST1 are aligned and coloured according to the ClustalW convention. The secondary structure is indicated with cylinders representing α-helices, solid lines representing structured loop regions, and dashed lines representing disordered regions. Grey bars (labelled ‘in’ and ‘out’) indicate approximate boundaries of transmembrane regions.
Extended Data Figure 2 Fab binding to BEST1cryst in the presence and absence of Ca2+.
The binding of the Fab to BEST1cryst was assayed by determining the amount of free Fab as a function of the concentration of BEST1cryst in the presence of either 10 μM Ca2+ or 5 mM EGTA (zero Ca2+) (Methods). The fraction of Fab bound is plotted with respect to the concentration of BEST1cryst. The curves correspond to fits of: fraction of Fab bound = [BEST1]h/(Kdh + [BEST1]h), where Kd is the equilibrium dissociation constant, h is the Hill coefficient, and [BEST1] is the BEST1cryst concentration. Derived parameters are: Kd = 15 nM in the presence of Ca2+ (h = 1.3) and Kd = 350 nM in the absence of Ca2+ (h = 1.3).
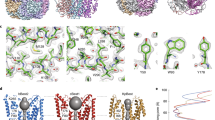
Extended Data Figure 3 Electron density and the C-terminal tail.
a, 2Fo − Fc electron density is shown, in stereo, for an area surrounding one of the five identical Ca2+ binding sites. The density was calculated from 40 to 2.85 Å resolution and contoured at 1.5σ (blue mesh) and 7σ (orange mesh) in the context of the final atomic model, which is shown as sticks and spheres (cyan sphere, calcium; red sphere, water). b, Electron density for the C-terminal tail. 2Fo − Fc electron density (blue mesh, calculated from 40 to 2.85 Å, and contoured at 1.5σ) is shown for the C-terminal tail of the yellow coloured subunit. c, Expanded view highlighting the electron density near Ser 358. Consistent with the electron density, mass spectrometry analysis of tryptic peptides of purified BEST1cryst detected only peptides containing Ser 358 that were not phosphorylated (Supplementary Discussion).
Extended Data Figure 4 Overall structures of the BEST1cryst–Fab complex.
a, Structure of the BEST1cryst–Fab complex in the P21 crystal form, viewed from the extracellular side. Fab molecules are grey and BEST1 subunits are coloured individually with α-helices depicted as cylinders. b, Orthogonal view showing approximate boundaries of the membrane. For clarity, two Fabs are drawn. c, C2 crystal form. Overall structures of the two BEST1cryst–Fab complexes in the asymmetric unit of the C2 crystal form are depicted in cartoon representation. BEST1 subunits are coloured individually and Fabs are grey.
Extended Data Figure 5 Ca2+-dependent activation of Best1cryst and permeability of the BEST1cryst–Fab complex.
a, Schematic of the fluorescence-based flux assay. Vesicles diluted into various test salts establish ion gradients. Anion influx through BEST1 produces a negative electric potential within the liposomes that drives the uptake of protons through an ionophore (CCCP) and quenches the fluorescence of a pH indicator (ACMA). b, Ca2+-dependent activation of BEST1cryst using NO3− as the permeant anion. The experimental setup was identical to that for Fig. 1a, except that NO3− was used as the permeant ion. Data presented here and in Fig. 1a were collected on the same day using the same batch of proteoliposomes and indicate the higher permeability of NO3− relative to Cl−. Free concentrations of Ca2+ are indicated. c, Ionic permeability of the BEST1cryst–Fab complex. The experiment setup is identical to that shown in Fig. 1b, except that it was performed using proteoliposomes reconstituted with the BEST1cryst–Fab complex. The Fab remained bound to the channel following reconstitution and excess Fab was maintained throughout (Methods). The slight differences in the shape of the curves for the BEST1cryst and BEST1cryst–Fab samples (for example, the lower rate of fluorescence decrease for Cl– compared with Fig. 1b) are in accord with variability observed among different liposome preparations.
Extended Data Figure 6 Molecular surface, subunit topology and anion binding in the outer entryway.
a, The molecular surface of the channel is shown in the same orientation as Fig. 2a and coloured according to electrostatic potential (red, −10 kT e−1; grey, neutral; blue, +10 kT e−1). An asterisk marks the location of the acidic cluster in the foreground. Approximate boundaries for the membrane are indicated. b, Subunit topology. N-terminal ends of α-helices exposed to the pore are indicated by +. The colouring corresponds to that of Fig. 2b. c, Anion binding in the outer entryway. Extracellular cut-away view of the molecular surface of BEST1 (orthogonal representation of Fig. 4a), revealing the surface of the pore (coloured by electrostatic potential; red, −10 kT e−1; white, neutral; blue, +10 kT e−1) and anomalous difference electron density for Br− ions (magenta mesh; 45–5 Å, non-crystallographic symmetry averaged, 8σ contour) in sites 1 and 2.
Extended Data Figure 7 Geometry within the neck and the possibility of anion–π interactions.
a, b, Representations of the pore at Phe 80 (a) and Phe 84 (b) are shown as sticks. The distance (d) from the central axis of the pore (black sphere) to the centre of the face of the aromatic ring is shown. An angle θ is defined as the angle between this distance vector and the plane of the ring. The geometry indicated corresponds to the crystal obtained in cymal-6. For the cymal-6-NG crystal, the values are: d = 3.9 Å, θ = 45° (Phe 80) and d = 4.8 Å, θ = 44° (Phe 84). c, Space-filling CPK representation of the pore at Phe 80, showing a hypothetical Cl− (green) positioned in the centre. Standard radii were used for the figure (carbon = 1.7 Å; Cl− = 1.81 Å). δ+ and – represent partial charges on the edge of the aromatic rings and the charge on Cl−, respectively.
Extended Data Figure 8 Evidence for coupling between the Ca2+ clasp and the gate from crystals grown in different detergents.
Comparison among crystals grown using different detergents gives insight into the channel’s gate and it’s coupling to Ca2+. Well-diffracting crystals belonging to the P21 space group were obtained using either the detergent cymal-6 or the detergent cymal-6-NG. Electron density maps indicated the presence of ordered cymal-6-NG but not cymal-6 molecules bound to the S1a–S1b components of the Ca2+ clasps (a). In addition, difference Fourier electron density maps suggested a slight widening of the neck of the pore in the structure with cymal-6 (b). Accordingly, while refined structures superimpose with an overall root mean squared deviation of only 0.15 Å, the diameter of the pore in the hydrophobic neck is ∼0.5 Å wider at Phe 80 for crystals in cymal-6 than it is with cymal-6-NG. Differences on the order of 0.3 Å between the atomic models are localized to the region near the Ca2+ clasp and to the neck of the pore (a). The subtle effects are an indication that changes in or around the Ca2+ clasp induce changes in the neck of the pore and they may hint at the mechanism of gating. a, 2FO − FC electron density for cymal-6-NG detergent molecules, contoured at 1.2σ, is shown as blue mesh in the context of the channel. The channel, with α-helices depicted as cylinders, is coloured on a yellow-to-red spectrum according to the displacement of Cα atoms between the refined atomic models obtained from crystals grown in cymal-6 and cymal-6-NG. Yellow colour represents displacements less than 0.15 Å and red colour represents displacements greater than 0.3 Å. An arrow indicates the neck of the pore and teal spheres denote Ca2+. b, Conformational shift in the gate. Phe 80 and surrounding residues of the refined structures from crystals in cymal-6 and cymal-6-NG are shown as sticks (coloured cyan and yellow, respectively) and viewed along the channel’s axis of symmetry from the extracellular side. Superimposed on this is an Fcymal-6 − Fcymal-6-NG difference Fourier map, which is calculated from 25 Å to 3.5 Å resolution and contoured at −3.8σ (magenta mesh) and +3.8σ (blue mesh).
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and additional references. (PDF 198 kb)
Rights and permissions
About this article
Cite this article
Kane Dickson, V., Pedi, L. & Long, S. Structure and insights into the function of a Ca2+-activated Cl− channel. Nature 516, 213–218 (2014). https://doi.org/10.1038/nature13913
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13913
This article is cited by
-
Cryo-EM structures of pannexin 1 and 3 reveal differences among pannexin isoforms
Nature Communications (2024)
-
Structures and gating mechanisms of human bestrophin anion channels
Nature Communications (2022)
-
Role of ANO1 in tumors and tumor immunity
Journal of Cancer Research and Clinical Oncology (2022)
-
Rhodopsin-bestrophin fusion proteins from unicellular algae form gigantic pentameric ion channels
Nature Structural & Molecular Biology (2022)
-
Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.