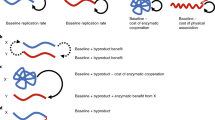
Abstract
Cooperation is central to the emergence of multicellular life; however, the means by which the earliest collectives (groups of cells) maintained integrity in the face of destructive cheating types is unclear. One idea posits cheats as a primitive germ line in a life cycle that facilitates collective reproduction. Here we describe an experiment in which simple cooperating lineages of bacteria were propagated under a selective regime that rewarded collective-level persistence. Collectives reproduced via life cycles that either embraced, or purged, cheating types. When embraced, the life cycle alternated between phenotypic states. Selection fostered inception of a developmental switch that underpinned the emergence of collectives whose fitness, during the course of evolution, became decoupled from the fitness of constituent cells. Such development and decoupling did not occur when groups reproduced via a cheat-purging regime. Our findings capture key events in the evolution of Darwinian individuality during the transition from single cells to multicellularity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bonner, J. T. The origins of multicellularity. Integr. Biol. 1, 27–36 (1998)
Okasha, S. Evolution and the Levels of Selection (Oxford Univ. Press, 2006)
Maynard Smith, J. & Szathmary, E. The Major Transitions in Evolution (Oxford Univ. Press, 1995)
Buss, L. W. The Evolution of Individuality (Princeton Univ. Press, 1987)
Godfrey-Smith, P. Darwinian Populations and Natural Selection (Oxford Univ. Press, 2009)
Nowak, M. A. Five rules for the evolution of cooperation. Science 314, 1560–1563 (2006)
Sachs, J. L., Mueller, U. G., Wilcox, T. P. & Bull, J. J. The evolution of cooperation. Q. Rev. Biol. 79, 135–160 (2004)
Rainey, P. B. & De Monte, S. Resolving conflicts during the evolutionary transition to multicellular life. Annu. Rev. Ecol. Evol. Syst. 45, 599–620 (2014)
Rainey, P. B. & Rainey, K. Evolution of cooperation and conflict in experimental bacterial populations. Nature 425, 72–74 (2003)
Pfeiffer, T., Schuster, S. & Bonhoeffer, S. Cooperation and competition in the evolution of ATP-producing pathways. Science 292, 504–507 (2001)
Maynard Smith, J. in Evolutionary Progress (ed. Nitecki, M. H. ) 219–230 (Univ. Chicago Press, 1988)
Sober, E. & Wilson, D. S. Unto Others: The Evolution and Psychology of Unselfish Behaviour (Harvard Univ. Press, 1998)
Velicer, G. J., Kroos, L. & Lenski, R. E. Developmental cheating in the social bacterium Myxococcus xanthus. Nature 404, 598–601 (2000)
Strassmann, J. E., Zhu, Y. & Queller, D. C. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature 408, 965–967 (2000)
Michod, R. E. Cooperation and conflict in the evolution of individuality. 2. Conflict mediation. Proc. R. Soc. Lond. B 263, 813–822 (1996)
Nunney, L. Group selection, altruism, and structured-deme models. Am. Nat. 126, 212–230 (1985)
Wade, M. J. & Breden, F. The evolution of cheating and selfish behavior. Behav. Ecol. Sociobiol. 7, 167–172 (1980)
Santorelli, L. A. et al. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature 451, 1107–1110 (2008)
Michod, R. E. Darwinian Dynamics: Evolutionary Transitions in Fitness and Individuality (Princeton Univ. Press, 1999)
Frank, S. A. Mutual policing and repression of competition in the evolution of cooperative groups. Nature 377, 520–522 (1995)
Queller, D. C. Relatedness and the fraternal major transitions. Phil. Trans. R. Soc. Lond. B 355, 1647–1655 (2000)
Rainey, P. B. Unity from conflict. Nature 446, 616 (2007)
McDonald, M. J., Gehrig, S. M., Meintjes, P. L., Zhang, X. X. & Rainey, P. B. Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics 183, 1041–1053 (2009)
Rainey, P. B. & Travisano, M. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72 (1998)
Bantinaki, E. et al. Adaptive divergence in experimental populations of Pseudomonas fluorescens. III. Mutational origins of wrinkly spreader diversity. Genetics 176, 441–453 (2007)
Goymer, P. et al. Adaptive divergence in experimental populations of Pseudomonas fluorescens. II. Role of the GGDEF regulator WspR in evolution and development of the wrinkly spreader phenotype. Genetics 173, 515–526 (2006)
Spiers, A. J., Kahn, S. G., Bohannon, J., Travisano, M. & Rainey, P. B. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161, 33–46 (2002)
Tarnita, C. E., Taubes, C. H. & Nowak, M. A. Evolutionary construction by staying together and coming together. J. Theor. Biol. 320, 10–22 (2013)
Trivers, R. L. The evolution of reciprocal altruism. Q. Rev. Biol. 46, 35–57 (1971)
Rainey, P. B. & Kerr, B. Cheats as first propagules: a new hypothesis for the evolution of individuality during the transition from single cells to multicellularity. Bioessays 32, 872–880 (2010)
Libby, E. & Rainey, P. B. A conceptual framework for the evolutionary origins of multicellularity. Phys. Biol. 10, 035001 (2013)
Libby, E. & Rainey, P. B. Eco-evolutionary feedback and the tuning of proto-developmental life cycles. PLoS ONE 8, e82274 (2013)
Lewontin, R. C. The units of selection. Annu. Rev. Ecol. Syst. 1, 1–18 (1970)
Wolpert, L. & Szathmary, E. Multicellularity: evolution and the egg. Nature 420, 745 (2002)
Leigh, E. G. How does selection reconcile individual advantage with the good of the group? Proc. Natl Acad. Sci. USA 74, 4542–4546 (1977)
Nunney, L. Lineage selection and the evolution of multistage carcinogenesis. Proc. R. Soc. Lond. B 266, 493–498 (1999)
Damuth, J. & Heisler, I. L. Alternative formulations of multi-level selection. Biol. Philos. 3, 407–430 (1988)
Michod, R. E. & Roze, D. in Mathematical and Computational Biology: Computational Morphogenesis, Hierarchical Complexity, and Digital Evolution (ed. Nehaniv, C. L. ) 47–92 (American Mathematical Society, 1999)
Okasha, S. Emergence, hierarchy and top-down causation in evolutionary biology. Interface Focus 2, 49–54 (2012)
De, N. et al. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6, e67 (2008)
Levinson, G. & Gutman, G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4, 203–221 (1987)
Moxon, E. R., Rainey, P. B., Nowak, M. A. & Lenski, R. E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4, 24–33 (1994)
Heilbron, K., Toll-Riera, M., Kojadinovic, M. & MacLean, R. C. Fitness is strongly influenced by rare mutations of large effect in a microbial mutation accumulation experiment. Genetics 197, 981–990 (2014)
Griesemer, J. The units of evolutionary transition. Selection 1, 67–80 (2001)
Wolpert, L. The evolution of development. Biol. J. Linn. Soc. 39, 109–124 (1990)
Silby, M. W. et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 10, R51 (2009)
Zhang, X. X. & Rainey, P. B. Construction and validation of a neutrally-marked strain of Pseudomonas fluorescens SBW25. J. Microbiol. Methods 71, 78–81 (2007)
Acknowledgements
We thank S. Nestmann for assistance with molecular aspects of the work and for guiding construction of the mutS deletion mutant. We thank E. Libby and Y. Pichugin for discussion, and S. De Monte and P. G. Smith for comments on drafts of the manuscript. We are indebted to PacBio and particularly J. Korlach and Y. Song for genome sequencing. P.B.R. currently holds an International Blaise Pascal Research Chair funded by the French State and the Ile-de-France, managed by the Fondation de l'Ecole Normale Supérieure. The work was directly supported by the Marsden Fund Council from government funding administered by the Royal Society of New Zealand, and in part by grant RFP-12-20 from the Foundational Questions in Evolutionary Biology Fund, by the National Science Foundation under Cooperative Agreement Number DBI-0939454, and by an NSF CAREER Award Grant (DEB0952825).
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and design of the study. K.H. and C.J.R. performed research, undertook data analysis and prepared figures. All authors wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Representative WS genotypes and fitness assays.
a, To analyse the response of derived lines to selection, a single representative WS genotype was obtained from each replicate population (set of eight microcosms) from both ancestral and derived lines. To obtain the representative set of derived types under the CE regime, SM colonies were collected from the end of phase I at generation 10 and pooled. The pooled sample was used to found phase II, at the end of which a single WS-type representative of each replicate was selected as described for the baseline (see below). This yielded 14 such types, one representing each replicate. To obtain the set of baseline types representing the ancestral state, SBW25 was used to found phase II. At the end of the 3-day period lines were harvested and plated. The single most abundant WS type from the most densely populated plate was selected as representative of that replicate. A third set of representative WS genotypes was obtained from lines evolved under the CP regime. This was as for the CE regime, but instead of pooling SM at the end of phase I, WS were collected and pooled. A single dominant WS type was chosen from each replicate. b, Cell- and line-fitness assays. Lines founded by representative WS genotypes (from ancestral and derived lineages) were competed against a marked (blue colonies) reference strain (SM and WS, for the CE and CP regimes, respectively). Use of the marked reference strain allowed the competitive performance of all ancestral and derived types to be assessed against a single common genotype. Cell fitness is the total number of cells in the mat after phase I, whereas line fitness is the proportion of evolved ‘offspring’ mats relative to a marked reference strain. In total 2,472 microcosms were assayed (three replicate assays per line).
Extended Data Figure 2 Line fitness and life cycle perpetuation.
a, b, Fitness of ancestral (a) and derived (b) lines and relationship with capacity to perpetuate the two-phase life cycle. Data are a breakdown of data in Fig. 3a. Colour intensity represents the proportion of three replicate microcosms harbouring the new type (white, absence of new type; dark red (yellow), presence of SM (WS) in all microcosms). Lines are ordered according to their fitness as assayed under the CE regime. Grey cells indicate loss of lines due to extinction.
Extended Data Figure 3 Life history traits under the CE regime.
a, Cell density of the new type. b, Proportion of the new cell type divided by the total number of cells. Each circle represents the mean of 42–45 lines (that is, three replicates for each of the 15 ancestral and 14 derived lines). Lines that failed to produce the required type were excluded. Black, derived; grey, ancestral. Error bars are s.e.m., based on n ≤ 15. *P < 0.05, using analysis of variance (ANOVA) and post hoc contrasts.
Extended Data Figure 4 Growth rate of SM.
Growth rate of the ancestral (ANC) and derived SM types from the CE and CP regimes obtained from the representative genotypes (biological and technical replicates; see Methods). (ANC, n = 81; CE, n = 95; CP n = 81). Error bars are s.e.m., based on n ≤ 15.
Extended Data Figure 5 Relationship between fitness and associated traits.
a–d, Summary of parameters describing line and cellular properties and the relationship among these parameters in ancestral (a) and derived (b) CE and ancestral (c) and derived (d) CP populations. Traits in the evolved populations (b, d) are depicted relative to their respective ancestral states (a, c): significant increase (large red circle), significant decrease (small blue circle), and no significant change (grey circle) using a generalized linear model (error structure: binomial; link function: logit) for line fitness and WS→SM with post hoc contrasts; and analysis of variance (ANOVA) for number of cells, number of WS, number of SM, and SM growth rate with post hoc contrasts. WS→SM, proportion of lines producing SM during phase I. Arrows indicate significant regressions and lines indicate significant correlations between traits, dashed lines indicate trends (0.05 < P < 0.09). The colour represents the direction of the relationship: red, positive; blue, negative. The significance level is P < 0.05 using Pearson and Spearman rank correlations, and regressions (line level fitness: generalized linear models; cell-level fitness and number of SM per μl (if present): general linear models). Individual cell properties displayed in a and c are identical for the ancestral state in both CE and CP regimes, but measures of line fitness are regime-specific, and transform the associations between parameters. Parameters that relate positively to line fitness in the CE regime negatively affect line fitness in the CP regime, and vice versa (a versus c). For example, in the CE regime, the number of SM cells and the rate at which WS cells give rise to SM cells positively regresses on line fitness (red arrows, a), whereas only the number of WS cells shows a positive regression with line fitness in the CP regime (red arrows, c). The relationships between parameters in the ancestral populations predict their evolutionary trajectory in each regime. After 10 generations of line selection the relationships between cell- and line-level parameters significantly altered in both CE and CP regimes (b and d). Line fitness improved in both regimes, thereby imposing selection on parameters that were linked to line fitness in their respective baselines. In the CE regime enhanced line fitness is explained by a significant increase in the capacity to transition from WS to SM and is not explained by enhanced performance of single cells: the fitness of single cells either remained unaltered or declined. Increased line fitness can be seen as a product of selection at the higher (group) level. In marked contrast is the CP regime where improved line fitness is readily explained by changes in traits that improve the competitive ability of individual cells. Enhanced line fitness in the CP regime can be interpreted as a by-product of selection at the lower (cell) level.
Extended Data Figure 6 Cycling through phases.
Ancestral and derived lines differ in their capacity to transition between phases of the life cycle. Three replicate populations of the two ancestral and derived lineages with the highest fitness: 71, 73 and 16, 17, respectively, were founded by the representative WS type and plated to check for SM types. Whereas ancestral WS took 48 h to generate detectable levels of SM, the two derived WS populations contained a mixture of WS and SM colonies, such that even at the time of initial inoculation, both types were present. SM colonies were then used to found populations that were plated to check for WS types. Ancestral lineages 71 and 73 completed two cycles before extinction through failure to produce SM, whereas derived lines 16 and 17 completed five cycles before termination of the experiment. Colonies were photographed after 48 h of growth. Notable in the evolved lines is the visible presence of zones of SM cells surrounding the central WS colony.
Extended Data Figure 7 Mechanism of life cycle transition.
a, Outline of the evolutionary history of line 17 from its founding by ancestral SM1 SBW25 through ten generations of the life cycle, by which time a mutS mutation had arisen. At the eleventh generation, the mutS (A1489C) mutation in WS22 was reverted to wild type (mutSWT) by in vitro manipulation. The two WS22 lineages (with and without the mutS mutation) were taken through three additional ‘expedited’ life-cycle generations, although with a cycling period of 24 h for mutS (A1489C) and 48 h for mutSWT. Genome sequences were acquired from five different time points (SM1, WS8, WS22, SM27 and WS28, indicated by surrounding boxes). For details of mutations see Supplementary Table 1. b, The mutS-dependent genetic switch. Four independent cultures of the generation-11 WS (WS22), with and without the mutS (A1489C) mutation, were passaged through an additional three ‘expedited’ generations of the life cycle (WS22–WS28). The nucleotide sequence of wspR was determined at each stage. Depicted is the tract of guanine residues beginning at nucleotide 742: WspR is active (and the phenotype is WS) when the tract length is seven residues, but inactive (and the phenotype is SM) when the tract length is eight. Red arrows show slippage events resulting in the gain or loss of a single guanine residue. Transitioning between phases is more reliable (fewer extinction events) and more likely to occur via the tract of guanine residues in the presence of mutS (A1489C). Lines of mutS (A1489C) were passaged on a 24-h cycle; mutSWT lines were passaged on a 48-h cycle (on a 24-h cycle mutSWT lines were extinct before the first generation completed—ancestral lines are incapable of transitioning on a 48-h cycle). Extinction events occurred whenever the extant phase failed to produce the next stage in the life cycle; death of a line allowed birth of an extant lineage (diagonal arrows).
Extended Data Figure 8 Life history traits under the CP regime.
a, Proportion of lines producing SM. b, Total cell density. c, Cell density of the new type. d, Proportion of SM cell types divided by the total number of cells. Each square represents the mean of 45 lines (that is, 3 replicates for each of the 15 lines); however, for c and d lines that failed to produce SM were excluded. Black, derived; grey, ancestral. Error bars are s.e.m., based on n ≤ 15. *P < 0.05, using a generalized linear model (error structure: binomial; link function: logit) and post hoc contrasts for a; and using analysis of variance (ANOVA) and post hoc contrasts for b–d.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary References and Supplementary Table 1. (PDF 1415 kb)
Rights and permissions
About this article
Cite this article
Hammerschmidt, K., Rose, C., Kerr, B. et al. Life cycles, fitness decoupling and the evolution of multicellularity. Nature 515, 75–79 (2014). https://doi.org/10.1038/nature13884
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13884
This article is cited by
-
The role of the ecological scaffold in the origin and maintenance of whole-group trait altruism in microbial populations
BMC Ecology and Evolution (2023)
-
Bacteria evolve macroscopic multicellularity by the genetic assimilation of phenotypically plastic cell clustering
Nature Communications (2023)
-
Scaffolds and scaffolding: an explanatory strategy in evolutionary biology
Biology & Philosophy (2023)
-
A coarse-graining account of individuality: how the emergence of individuals represents a summary of lower-level evolutionary processes
Biology & Philosophy (2023)
-
Moving Past Conventionalism About Multilevel Selection
Erkenntnis (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.