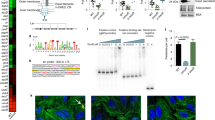
Abstract
Systemic infection induces conserved physiological responses that include both resistance and ‘tolerance of infection’ mechanisms1. Temporary anorexia associated with an infection is often beneficial2,3, reallocating energy from food foraging towards resistance to infection4 or depriving pathogens of nutrients5. However, it imposes a stress on intestinal commensals, as they also experience reduced substrate availability; this affects host fitness owing to the loss of caloric intake and colonization resistance (protection from additional infections)6. We hypothesized that the host might utilize internal resources to support the gut microbiota during the acute phase of the disease. Here we show that systemic exposure to Toll-like receptor (TLR) ligands causes rapid α(1,2)-fucosylation of small intestine epithelial cells (IECs) in mice, which requires the sensing of TLR agonists, as well as the production of interleukin (IL)-23 by dendritic cells, activation of innate lymphoid cells and expression of fucosyltransferase 2 (Fut2) by IL-22-stimulated IECs. Fucosylated proteins are shed into the lumen and fucose is liberated and metabolized by the gut microbiota, as shown by reporter bacteria and community-wide analysis of microbial gene expression. Fucose affects the expression of microbial metabolic pathways and reduces the expression of bacterial virulence genes. It also improves host tolerance of the mild pathogen Citrobacter rodentium. Thus, rapid IEC fucosylation appears to be a protective mechanism that utilizes the host’s resources to maintain host–microbial interactions during pathogen-induced stress.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ayres, J. S. & Schneider, D. S. Tolerance of infections. Annu. Rev. Immunol. 30, 271–294 (2012)
Ayres, J. S. & Schneider, D. S. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7, e1000150 (2009)
Murray, M. J. & Murray, A. B. Anorexia of infection as a mechanism of host defense. Am. J. Clin. Nutr. 32, 593–596 (1979)
Kyriazakis, I. I., Tolkamp, B. J. & Hutchings, M. R. Towards a functional explanation for the occurrence of anorexia during parasitic infections. Anim. Behav. 56, 265–274 (1998)
Exton, M. S. Infection-induced anorexia: active host defence strategy. Appetite 29, 369–383 (1997)
Stecher, B. & Hardt, W. D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 14, 82–91 (2011)
Bocci, V. & Winzler, R. J. Metabolism of l-fucose-1–14C and of fucose glycoproteins in the rat. Am. J. Physiol. 216, 1337–1342 (1969)
Becker, D. J. & Lowe, J. B. Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R (2003)
Kashyap, P. C. et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl Acad. Sci. USA 110, 17059–17064 (2013)
Ng, K. M. et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013)
Umesaki, Y., Tohyama, K. & Mutai, M. Appearance of fucolipid after conventionalization of germ-free mice. J. Biochem. 90, 559–561 (1981)
Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. I. A model of host-microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383 (1996)
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005)
Clark, M. A., Jepson, M. A., Simmons, N. L., Booth, T. A. & Hirst, B. H. Differential expression of lectin-binding sites defines mouse intestinal M-cells. J. Histochem. Cytochem. 41, 1679–1687 (1993)
Kinnebrew, M. A. et al. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36, 276–287 (2012)
Thomsson, K. A. et al. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fuca1-2 glycosyltransferase. Biochem. J. 367, 609–616 (2002)
Holmén, J. M., Olson, F. J., Karlsson, H. & Hansson, G. C. Two glycosylation alterations of mouse intestinal mucins due to infection caused by the parasite Nippostrongylus brasiliensis. Glycoconj. J. 19, 67–75 (2002)
Domino, S. E., Zhang, L. & Lowe, J. B. Molecular cloning, genomic mapping, and expression of two secretor blood group α (1,2)fucosyltransferase genes differentially regulated in mouse uterine epithelium and gastrointestinal tract. J. Biol. Chem. 276, 23748–23756 (2001)
Domino, S. E., Zhang, L., Gillespie, P. J., Saunders, T. L. & Lowe, J. B. Deficiency of reproductive tract α(1,2)fucosylated glycans and normal fertility in mice with targeted deletions of the FUT1 or FUT2 α(1,2)fucosyltransferase locus. Mol. Cell. Biol. 21, 8336–8345 (2001)
Vaishnava, S. et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011)
Hovel-Miner, G., Faucher, S. P., Charpentier, X. & Shuman, H. A. ArgR-regulated genes are derepressed in the Legionella-containing vacuole. J. Bacteriol. 192, 4504–4516 (2010)
Zhang, Z., Yen, M. R. & Saier, M. H., Jr Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli. J. Bacteriol. 192, 2013–2019 (2010)
Fischbach, M. A. & Sonnenburg, J. L. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10, 336–347 (2011)
Kamada, N. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012)
Barba, J. et al. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187, 7918–7930 (2005)
Scott, K. P., Martin, J. C., Campbell, G., Mayer, C. D. & Flint, H. J. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. J. Bacteriol. 188, 4340–4349 (2006)
Pacheco, A. R. et al. Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117 (2012)
De Vadder, F. et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96 (2014)
McGovern, D. P. et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum. Mol. Genet. 19, 3468–3476 (2010)
Morrow, A. L. et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. J. Pediatr. 158, 745–751 (2011)
Kleinridders, A. et al. MyD88 signaling in the CNS is required for development of fatty acid-induced leptin resistance and diet-induced obesity. Cell Metab. 10, 249–259 (2009)
Eberl, G. et al. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nature Immunol. 5, 64–73 (2004)
Cua, D. J. et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748 (2003)
Zenewicz, L. A. et al. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity 27, 647–659 (2007)
Malick, L. E. & Wilson, R. B. Modified thiocarbohydrazide procedure for scanning electron microscopy: routine use for normal, pathological, or experimental tissues. Stain Technol. 50, 265–269 (1975)
Sakamoto, M. & Ohkuma, M. Identification and classification of the genus Bacteroides by multilocus sequence analysis. Microbiology 157, 3388–3397 (2011)
Boronat, A. & Aguilar, J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140, 320–326 (1979)
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675 (2012)
Ubeda, C. et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341 (2010)
Buffie, C. G. et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80, 62–73 (2012)
Fleming, S. E., Traitler, H. & Koellreuter, B. Analysis of volatile fatty acids in biological specimens by capillary column gas chromatography. Lipids 22, 195–200 (1987)
Tangerman, A. & Nagengast, F. M. A gas chromatographic analysis of fecal short-chain fatty acids, using the direct injection method. Anal. Biochem. 236, 1–8 (1996)
Zhao, G., Nyman, M. & Jonsson, J. A. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed. Chromatogr. 20, 674–682 (2006)
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336 (2010)
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013)
Chirgwin, J. M., Przybyla, A. E., MacDonald, R. J. & Rutter, W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18, 5294–5299 (1979)
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012)
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA 108 (suppl. 1). 4516–4522 (2011)
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006)
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011)
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280 (2004)
Ning, Z., Cox, A. J. & Mullikin, J. C. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001)
Abubucker, S. et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358 (2012)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Acknowledgements
We thank C. Reardon and C. Daly for sequencing support, H. Ye for help with metabolic cage analysis, N. F. Dalleska for assistance and use of GC–MS instrumentation in the Environmental Analysis Center at the California Institute of Technology, and G. Nuñez for luciferase-expressing C. rodentium. This work was supported by grants from the National Institutes of Health (P50 GM068763 to P.J.T., AI96706 and AI42135 to E.G.P., T32 AI065382 to J.M.P.), the Harvard Bauer Fellows Program, National Science Foundation grant EFRI-1137089 to R.F.I. and A.V.C., Digestive Disease Research Core Center grant DK42086 and a Kenneth Rainin Foundation grant to A.V.C.
Author information
Authors and Affiliations
Contributions
J.M.P., M.A.K., M.C.A. and E.G.P. performed analysis of inducible fucosylation in mice, including mutant strains; J.M.P. and C.F.M. produced DNA and RNA sequencing data and P.J.T. analysed these data; D.S. produced Myd88fl/fl mice; T.V.G. produced GF BALB/c mice and performed cytokine ELISA analysis; S.R.B. and R.F.I. performed analysis of short-chain fatty acids; R.F.I., E.G.P. and P.J.T. contributed to writing of the manuscript; A.V.C. conceived the project, analysed the results and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Requirements and kinetics for small intestine fucosylation induced by systemic injection of TLR ligands.
a, Systemic injection of bacterial TLR ligands induces small intestine fucosylation, but simple starvation does not. UEA-1 staining (as in Fig. 1) after i.p. injection of CpG DNA, or Pam3CSK4, or food deprivation for 24 h of BALB/c SPF mouse. b, LPS injection causes small intestine fucosylation in various inbred mouse strains. SPF mice of the indicated strains were injected with LPS i.p. and the small intestine was stained with UEA-1 after 24 h, as in Fig. 1. c, Fucosylation peaks at 8 h after LPS injection and is still detectable at 96 h. d, M cells can be readily detected by scanning electron microscope and UEA-1 staining of the domes of the Peyer’s patches, but are rare in the villi and are not massively induced in the villi by LPS injection. UEA-1 staining and scanning electron microscopy were performed on adjacent pieces from the proximal one-third of the small intestine. Scale bars = 100 μm for UEA-1 staining, 50 μm for scanning electron microscope images. e, Small intestine fucosylation does not require the presence of endogenous microbiota (LPS injection in GF mouse) and is not induced by oral administration of LPS (1 mg). All data are representative of at least two independent experiments.
Extended Data Figure 2 MyD88-dependent pathway for fucosylation of small intestine IECs in response to systemic stimulation of TLRs.
a, FACS analysis of IECs from three segments of small intestine from the indicated mice. Cells are gated on the FSC/SSC high epithelial cell population. At least two mice per mutant genotype were stained along with two control mice in the experiments shown. b, SPF mice were pre-treated with 20 mg streptomycin and orally infected with S. enterica Typhimurium. The small intestine was stained at 24 h post-infection. MyD88 expression was necessary in CD11c+ cells but not villin+ IECs for S. enterica Typhimurium-induced fucosylation. Data are representative of at least two independent experiments.
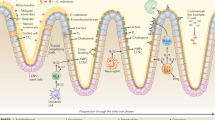
Extended Data Figure 3 A proposed model for the mechanisms linking inducible fucosylation to the gut microbiota.
Systemic microbial agonists activate TLRs on CD11c+ dendritic cells (DCs), causing secretion of the cytokine IL-23, which in turn stimulates RORγt-dependent ILCs to secrete IL-22. IL-22 causes small intestine epithelial cells to upregulate Fut2. Fucosylated proteins are either secreted into the lumen or expressed on the cell surface and later shed into the lumen. Fucosidase-expressing bacteria (blue) liberate fucose residues, which they can utilize and share with other bacteria lacking the fucose-cleaving enzyme. Bacterial metabolism of fucose potentially produces metabolites such as short-chain fatty acids (SCFAs). Fucose also directly or indirectly downregulates virulence gene expression by pathobionts (red) or bona fide pathogens27.
Extended Data Figure 4 Consequences of LPS injection in Fut2-sufficient and Fut2-deficient BALB/c mice.
a, Inflammatory cytokines IL-1β, IL-6 and TNF-α were measured by ELISA in sera of mice before or 2 h after injection with LPS (4 h for IL-1β). abx, mice on antibiotic water for 2 days before injection. Bars are mean ± s.e.m.; ND, not detected. Data are combined from three experiments. b, Expression of RegIIIγ (also regulated by the MyD88–IL23–IL22 pathway). Measurement by qPCR of reg3g gene expression in mid-small-intestine tissue, relative to gapdh (ddCt method). Numbers indicate mean fold change ± s.e.m. in LPS-treated versus untreated mice. Differences between LPS-treated Fut2+ and abx or Fut2− levels are not significant (P > 0.05, two-tailed Student’s t-test). Data are combined from three experiments. c, Weight loss and recovery is not different in Fut2+/− and Fut2−/− mice after simple starvation (mean ± s.e.m., P > 0.05 at all time points, two-tailed Student’s t-test; NS, not significant). d, Lack of direct toxic effect of antibiotics (abx) measured as the weight loss of BALB/c GF animals treated with LPS i.p (mean ± s.e.m., P > 0.05 by two-tailed Student’s t-test at all time points). Data are combined from two experiments. e–g, Similar total bacterial loads in Fut2+/− and Fut2−/− mice before and after LPS injection and antibiotic treatment. Total bacterial loads in faeces were estimated by plating on aerobic (e) and anaerobic (f) non-selective media, and by qPCR for 16S gene copies (g). There were no significant differences between Fut2-sufficient (filled circles) and Fut2-deficient (open circles) mice before or after LPS treatment (two-tailed Student’s t-test). Circles indicate individual mice; horizontal lines indicate means; red circles indicate antibiotic-treated mice. Data are combined from three experiments.
Extended Data Figure 5 Fucosylated protein in IECs and gut contents.
a, Proteins α(1,2)fucosylated in IECs after LPS injection identified by UEA-1 precipitation and mass spectrometry. Abundance is the number of peptide fragments attributed to each gene. b, IECs from Fut2+ untreated, Fut2+ LPS-treated, or Fut2− untreated mice were isolated, and lysates separated by SDS–PAGE. α(1,2)fucosylated proteins were detected by blotting with UEA-1 lectin conjugated to horseradish peroxidase (HRP). c, Identical gel stained with Coomassie blue for total protein content. d, Relative density of the boxed area of each lane from b divided by the relative density in c. e, UEA-1 staining of luminal proteins as in Fig. 3c. Blot is overexposed to show absence of luminal fucosylated proteins in the LPS-treated, Fut2− mouse. b–e, Data are representative of two independent experiments.
Extended Data Figure 6 Generation of fucose-sensing reporter bacterial strains.
a, Reporter E. coli were grown to stationary phase in minimal medium containing 10 mM glucose and the indicated concentrations of l-fucose (asterisk indicates promoter-less vector), and GFP fluorescence was measured. b, Fucosidase activity is dramatically reduced after 2 days of antibiotics (abx) treatment but recovers after cessation of treatment. Measurement of total α-l-fucosidase activity in faeces. Faecal supernatant was assayed for cleavage of 4-methylumbelliferyl-fucopyranoside substrate by fluorescence. n = 5 SPF antibiotics-treated, 3 GF mice. c, Faecal homogenates were plated anaerobically on BHIS agar containing 5-bromo-4-chloro-3-indolyl α-l-fucopyranoside, which forms a blue precipitate upon cleavage of the fucosyl residue. Both blue and white colonies are present. d, Pure cultures of Bacteroides species were streaked on the same medium as in c. B. uniformis (left) is not predicted to carry an α-l-fucosidase gene, and remains white; B. acidifaciens (middle) and B. thetaiotaomicron (right) both express fucosidase activity and develop blue colonies. e, Loss of B. acidifaciens from the faeces of mice treated with antibiotics (Abx) in water (PCR for the gyrB gene). C−, water control; C+, B. acidifaciens genomic DNA. f, Summary of reporter E. coli experiments in SPF mice (representative experiment is shown in Fig. 3e). Points are mean GFP fluorescence from all reporter bacteria measured in each of three independent experiments (n = 65 bacteria per mouse; *P < 0.05, Student’s t test).
Extended Data Figure 7 Microbial community structure is impacted by cohousing yet robust to host fucosylation and LPS exposure, whereas microbial gene expression depends on Fut2.
a, Stable relative abundance of bacterial phyla across treatment groups and genotypes, as indicated by 16S rRNA gene sequencing. Values represent the mean abundance of phyla found at >1% relative abundance in at least one sample. b, Unweighted UniFrac analysis of the gut microbiota of Fut2-deficient (no outline) and Fut2-sufficient (black outline) mice. Points are coloured based on kinship and labelled by time point (before or after LPS exposure). Results are based on 180,000 randomly selected 16S rRNA gene sequences per sample. c, Microbial diversity as measured by the Shannon diversity index (n = 178,100 sequences per sample). Values are mean ± s.e.m. (n = 3 Fut2+, 4 Fut2− mice per time point). d, KEGG modules and pathways expressed in microbiota at higher levels after LPS exposure in Fut2-positive (left) and Fut2-negative mice (right) (n = 3 per group; Humann/LefSe analysis; LDA >2).
Extended Data Figure 8 Lack of indicible fucosylation and small intestine colonization in C. rodentium-infected mice.
a, C. rodentium causes no small intestine fucosylation in SPF mice at day (d)3, day 7, or day 12 post-infection (p.i.). b, Small intestine colonization by C. rodentium is low regardless of Fut2 expression and LPS treatment. Small intestine contents were removed by gentle squeezing, homogenized in PBS, and plated. Data are shown as mean ± s.e.m.; n = 4. NS, not significant. Dotted line shows the limit of detection. Data are representative of two experiments.
Extended Data Figure 9 Effect of exogenous fucose on caecal short-chain fatty acid levels.
Caecal short-chain fatty acids were measured after gavaging starved mice with the indicated sugars (100 mM concentration). Fucose gavage leads to increased propionate production in SPF but not GF mice. Data are shown as mean ± s.e.m. **P < 0.01, Student’s two-tailed t-test. ND, not detected. Data are combined from three experiments.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-2. (PDF 1094 kb)
Rights and permissions
About this article
Cite this article
Pickard, J., Maurice, C., Kinnebrew, M. et al. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature 514, 638–641 (2014). https://doi.org/10.1038/nature13823
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13823
This article is cited by
-
A non-human primate model for human norovirus infection
Nature Microbiology (2024)
-
FUT1-mediated terminal fucosylation acts as a new target to attenuate renal fibrosis
Molecular Medicine (2023)
-
Sub-1.4 cm3 capsule for detecting labile inflammatory biomarkers in situ
Nature (2023)
-
Innate lymphoid cells and cancer
Nature Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.