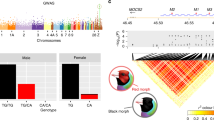
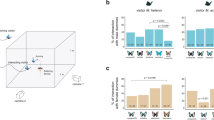
Abstract
The monarch butterfly, Danaus plexippus, is famous for its spectacular annual migration across North America, recent worldwide dispersal, and orange warning colouration. Despite decades of study and broad public interest, we know little about the genetic basis of these hallmark traits. Here we uncover the history of the monarch’s evolutionary origin and global dispersal, characterize the genes and pathways associated with migratory behaviour, and identify the discrete genetic basis of warning colouration by sequencing 101 Danaus genomes from around the globe. The results rewrite our understanding of this classic system, showing that D. plexippus was ancestrally migratory and dispersed out of North America to occupy its broad distribution. We find the strongest signatures of selection associated with migration centre on flight muscle function, resulting in greater flight efficiency among migratory monarchs, and that variation in monarch warning colouration is controlled by a single myosin gene not previously implicated in insect pigmentation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dingle, H., Zalucki, M. P., Rochester, W. A. & Armijo-Prewitt, T. Distribution of the monarch butterfly, Danaus plexippus (L.) (Lepidoptera: Nymphalidae), in western North America. Biol. J. Linn. Soc. 85, 491–500 (2005)
Lyons, J. I. et al. Lack of genetic differentiation between monarch butterflies with divergent migration destinations. Mol. Ecol. 21, 3433–3444 (2012)
Malcolm, S. B. & Zalucki, M. P. Biology and Conservation of the Monarch Butterfly (Natural History Museum of LA County, 1993)
Oberhauser, K. S. & Solensky, M. J. The Monarch Butterfly: Biology and Conservation (Cornell Univ. Press, 2004)
Urquhart, F. A. Found at last; the monarch’s winter home. Natl Geogr. Mag. 150, 161–173 (1976)
Urquhart, F. A. & Urquhart, N. R. Autumnal migration routes of the eastern population of the monarch butterfly (Danaus p. plexippus L.; Danaidae; Lepidoptera) in North America to the overwintering site in the Neovolcanic Plateau of Mexico. Can. J. Zool. 56, 1759–1764 (1978)
Wassenaar, L. I. & Hobson, K. A. Natal origins of migratory monarch butterflies at wintering colonies in Mexico: new isotopic evidence. Proc. Natl Acad. Sci. USA 95, 15436–15439 (1998)
Froy, O., Gotter, A. L., Casselman, A. L. & Reppert, S. M. Illuminating the circadian clock in monarch butterfly migration. Science 300, 1303–1305 (2003)
Heinze, S. & Reppert, S. M. Sun compass integration of skylight cues in migratory monarch butterflies. Neuron 69, 345–358 (2011)
Merlin, C., Gegear, R. J. & Reppert, S. M. Antennal circadian clocks coordinate sun compass orientation in migratory monarch butterflies. Science 325, 1700–1704 (2009)
Reppert, S. M., Gegear, R. J. & Merlin, C. Navigational mechanisms of migrating monarch butterflies. Trends Neurosci. 33, 399–406 (2010)
Sauman, I. et al. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron 46, 457–467 (2005)
Mouritsen, H. & Frost, B. J. Virtual migration in tethered flying monarch butterflies reveals their orientation mechanisms. Proc. Natl Acad. Sci. USA 99, 10162–10166 (2002)
Ackery, P. R. & Vane-Wright, R. I. Milkweed Butterflies: Their Cladistics and Biology (British Museum, 1984)
Altizer, S. & Davis, A. K. Populations of monarch butterflies with different migratory behaviors show divergence in wing morphology. Evolution 64, 1018–1028 (2010)
Dockx, C. Directional and stabilizing selection on wing size and shape in migrant and resident monarch butterflies, Danaus plexippus (L.), in Cuba. Biol. J. Linn. Soc. 92, 605–616 (2007)
Vane-Wright, R. I. in Biology and Conservation of the Monarch Butterfly (eds Malcolm, S. B. & Zalucki, M. P. ) 179–187 (Natural History Museum of LA County, 1993)
Haeger, J. F. & Jordano, D. The Monarch butterfly Danaus plexippus (Linnaeus, 1758) in the Strait of Gibraltar (Lepidoptera: Danaidae). SHILAP Rev. Lepidopterol. 37, 421–438 (2009)
Zhu, H., Casselman, A. & Reppert, S. M. Chasing migration genes: a brain expressed sequence tag resource for summer and migratory monarch butterflies (Danaus plexippus). PLoS ONE 3, e1345 (2008)
Zhan, S., Merlin, C., Boore, J. L. & Reppert, S. M. The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185 (2011)
Kitching, I. J., Ackery, P. R. & Vane-Wright, R. I. in Biology and Conservation of the Monarch Butterfly (eds Malcolm, S. B. & Zalucki, M. P. ) 11–16 (Natural History Museum of LA County, 1993)
Gauthreaux, S. A. in Avian Biology Vol. 4 (eds Farner, D. S., King, J. R. & Parkes, K. C. ) Ch. 2 93–168 (Elsevier, 1982)
Zalucki, M. P. & Clarke, A. R. Monarchs across the Pacific: the Columbus hypothesis revisited. Biol. J. Linn. Soc. 82, 111–121 (2004)
Brower, L. P., Oberhauser, K. S., Boppré, M., Brower, A. V. Z. & Vane-Wright, R. I. Monarch sex: ancient rites, or recent wrongs? Antenna 31, 12–18 (2007)
Peter, B. M. & Slatkin, M. Detecting range expansions from genetic data. Evolution 67, 3274–3289 (2013)
Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011)
Gutenkunst, R. N., Hernandez, R. D., Williamson, S. H. & Bustamante, C. D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 (2009)
Yi, X. et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010)
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010)
Schnorrer, F. et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464, 287–291 (2010)
Kelemen-Valkony, I. et al. Drosophila basement membrane collagen col4a1 mutations cause severe myopathy. Matrix Biol. 31, 29–37 (2012)
Plaisier, E. et al. COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps. N. Engl. J. Med. 357, 2687–2695 (2007)
Hakeda, S., Endo, S. & Saigo, K. Requirements of Kettin, a giant muscle protein highly conserved in overall structure in evolution, for normal muscle function, viability, and flight activity of Drosophila. J. Cell Biol. 148, 101–114 (2000)
Gibo, D. L. & Pallett, M. J. Soaring flight of monarch butterflies, Danaus plexippus (Lepidoptera: Danaidae), during the late summer migration in southern Ontario. Can. J. Zool. 57, 1393–1401 (1979)
Niitepõld, K. et al. Flight metabolic rate and Pgi genotype influence butterfly dispersal rate in the field. Ecology 90, 2223–2232 (2009)
Mitikka, V. & Hanski, I. Pgi genotype influences flight metabolism at the expanding range margin of the European map butterfly. Ann. Zool. Fenn. 47, 1–14 (2010)
Niitepõld, K., Mattila, A. L. K., Harrison, P. J. & Hanski, I. Flight metabolic rate has contrasting effects on dispersal in the two sexes of the Glanville fritillary butterfly. Oecologia 165, 847–854 (2011)
Reichstein, T., von Euw, J., Parsons, J. A. & Rothschild, M. Heart poisons in the monarch butterfly. Science 161, 861–866 (1968)
Ritland, D. B. & Brower, L. P. The viceroy butterfly is not a batesian mimic. Nature 350, 497–498 (1991)
Stimson, J. & Kasuya, M. Decline in the frequency of the white morph of the monarch butterfly (Danaus plexippus plexippus L. Nymphalidae) on Oahu, Hawaii. J. Lepid. Soc. 54, 29–32 (2000)
Stimson, J. S. & Meyers, L. Inheritance and frequency of a color polymorphism in Danaus plexippus (Lepidoptera: Danaidae) on Oahu, Hawaii. J. Res. Lepid. 23, 153–160 (1984)
Nijhout, H. F. The Development and Evolution of Butterfly Wing Patterns (Smithsonian Press, 1991)
Mercer, J. A., Seperack, P. K., Strobel, M. C., Copeland, N. G. & Jenkins, N. A. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature 349, 709–713 (1991)
Fukuda, M. & Kuroda, T. S. Missense mutations in the globular tail of myosin-Va in dilute mice partially impair binding of Slac2-a/melanophilin. J. Cell Sci. 117, 583–591 (2004)
Rendón-Salinas, E. & Tavera-Alonso, G. Forest Surface Occupied by Monarch Butterfly Hibernation Colonies in December 2013 (World Wildlife Fund-México, 2014)
Haber, W. A. in Biology and Conservation of the Monarch Butterfly (eds Malcolm, S. B. & Zalucki, M. P. ) 201–207 (Natural History Museum of LA County, 1993)
James, D. G. in Biology and Conservation of the Monarch Butterfly (eds Malcolm, S. B. & Zalucki, M. P. ) 189–200 (Natural History Museum of LA County, 1993)
Smith, D. A. S. & Owen, D. F. Colour genes as markers for migratory activity: The butterfly Danaus chrysippus in Africa. Oikos 78, 127–135 (1997)
Lunter, G. & Goodson, M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21, 936–939 (2011)
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 43, 491–498 (2011)
Zhan, S. & Reppert, S. M. MonarchBase: the monarch butterfly genome database. Nucleic Acids Res. 41, D758–D763 (2013)
Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98 (2012)
International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1036–1045 (2008)
She, R., Chu, J. S., Wang, K., Pei, J. & Chen, N. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 19, 143–149 (2009)
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004)
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007)
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010)
Xia, Q. et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326, 433–436 (2009)
PHYLIP. (phylogeny inference package) v. 3.6 http://evolution.genetics.washington.edu/phylip.html (Univ. Washington, 2005)
Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8, e1002967 (2012)
Tang, H., Peng, J., Wang, P. & Risch, N. J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 28, 289–301 (2005)
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006)
Knight, A. & Brower, L. P. The influence of eastern North American autumnal migrant monarch butterflies (Danaus plexippus L.) on continuously breeding resident monarch populations in southern Florida. J. Chem. Ecol. 35, 816–823 (2009)
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005)
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
Haag-Liautard, C. et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445, 82–85 (2007)
Keightley, P. D. et al. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19, 1195–1201 (2009)
Zalucki, M. P. & Clarke, A. R. Monarchs across the Pacific: the Columbus hypothesis revisited. Biol. J. Linn. Soc. 82, 111–121 (2004)
Nei, M. & Li, W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA 76, 5269–5273 (1979)
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989)
Durand, E. Y., Patterson, N., Reich, D. & Slatkin, M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010)
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013)
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009)
Hudson, R. R., Kreitman, M. & Aguade, M. A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159 (1987)
Murrell, B. et al. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol. Biol. Evol. 30, 1196–1205 (2013)
Murrell, B. et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 (2012)
Kosakovsky Pond, S. L. & Frost, S. D. W. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222 (2005)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 28, 511–515 (2010)
Bartholomew, G. A., Vleck, D. & Vleck, C. M. Instantaneous measurements of oxygen consumption during pre-flight warm-up and post-flight cooling in sphingid and saturniid moths. J. Exp. Biol. 90, 17–32 (1981)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012)
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Acknowledgements
We thank B. Ballister, S. Barribeau, R. Bartel, N. Chamberlain, R. Cook, A. Davis, D. Feary, D. Frey, M. Maudsley, G. Moreira, E. Osburn, R. Rarick, E. Rendon, D. Rodrigues, E. Sternberg and J. Stimson for assistance collecting or providing specimens. We also thank J. Jensen and D. Lohman for discussion. This work was supported by National Institutes of Health grant GM086794-02S1, National Science Foundation grants IOS-134367, DEB-0643831, DEB-1019746 and DEB-1316037, start-up funds from the Chinese Academy of Sciences and Shanghai Institutes for Biological Sciences, and Neubauer Funds from the University of Chicago.
Author information
Authors and Affiliations
Contributions
S.Z. designed and implemented analyses of dispersal and migration and co-wrote the manuscript. W.Z. performed wing colour analyses. K.N. performed respirometry experiments. J.H. helped design the project and collected and prepared samples for sequencing. J.F.H. and M.P.Z. provided samples and interpreted results. S.A., J.C.d.R. and S.M.R. helped design the project, provided samples, and interpreted results. M.R.K. conceived and directed the project, performed targeted population genetic analyses, and co-wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Relationships among monarch populations inferred using the maximum likelihood method implemented in Treemix.
Note, this is a fully resolved, bifurcating tree. The very short basal branches indicate little genetic drift in North American populations, not unresolved basal relationships. Colours correspond to those in Fig. 1. Treemix also inferred five migration events among populations: from Puerto Rico to Aruba, from Puerto Rico to Costa Rica, from New Caledonia to Fiji, from Belize or Costa Rica to Portugal, and from Belize to Puerto Rico.
Extended Data Figure 2 Demographic history of the monarch butterfly.
a, Patterns of linkage-disequilibrium decay across the genome in different geographic populations. b, Genome-wide distribution of minor allele frequencies. c, Heterozygosity across populations, estimated as the ratio of heterozygous SNPs to homozygous SNPs/individual. d, Demographic history inferred using PSMC. This analysis includes representative individuals of high sequencing depth for each geographic location. The period of the last glacial maximum (LGM; ∼20,000 years ago) is shaded in grey.
Extended Data Figure 3 ∂a∂i analysis parameter estimates.
a, Schematic of demographic scenario modelled in ∂a∂i labelled with parameters being estimated. Nu, effective population size (individuals); m, migration rate (individuals/year); T, time (years). b, Inferred parameter estimates. c, One-dimensional model-data comparison considering North America population only. In the left panel, the model is plotted in red and the data in blue. In the right panel, the residuals between model and data are plotted. d, Two-dimensional comparison for joint estimation of North America and dispersal populations (Central/South America, Pacific, Atlantic). The left two panels are marginal spectra for data and the maximum-likelihood model, respectively. The right two panels show the residuals.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-14. (PDF 422 kb)
Rights and permissions
About this article
Cite this article
Zhan, S., Zhang, W., Niitepõld, K. et al. The genetics of monarch butterfly migration and warning colouration. Nature 514, 317–321 (2014). https://doi.org/10.1038/nature13812
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13812
This article is cited by
-
Genome sequence of Ophryocystis elektroscirrha, an apicomplexan parasite of monarch butterflies: cryptic diversity and response to host-sequestered plant chemicals
BMC Genomics (2023)
-
Overwintering and breeding patterns of monarch butterflies (Danaus plexippus) in coastal plain habitats of the southeastern USA
Scientific Reports (2023)
-
Neural representation of goal direction in the monarch butterfly brain
Nature Communications (2023)
-
Lactobacillus for ribosome peptide editing cancer
Clinical and Translational Oncology (2023)
-
Comparative neurogenetics of dog behavior complements efforts towards human neuropsychiatric genetics
Human Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.