Abstract
The human immunodeficiency virus type 1 (HIV-1) envelope (Env) spike, comprising three gp120 and three gp41 subunits, is a conformational machine that facilitates HIV-1 entry by rearranging from a mature unliganded state, through receptor-bound intermediates, to a post-fusion state. As the sole viral antigen on the HIV-1 virion surface, Env is both the target of neutralizing antibodies and a focus of vaccine efforts. Here we report the structure at 3.5 Å resolution for an HIV-1 Env trimer captured in a mature closed state by antibodies PGT122 and 35O22. This structure reveals the pre-fusion conformation of gp41, indicates rearrangements needed for fusion activation, and defines parameters of immune evasion and immune recognition. Pre-fusion gp41 encircles amino- and carboxy-terminal strands of gp120 with four helices that form a membrane-proximal collar, fastened by insertion of a fusion peptide-proximal methionine into a gp41-tryptophan clasp. Spike rearrangements required for entry involve opening the clasp and expelling the termini. N-linked glycosylation and sequence-variable regions cover the pre-fusion closed spike; we used chronic cohorts to map the prevalence and location of effective HIV-1-neutralizing responses, which were distinguished by their recognition of N-linked glycan and tolerance for epitope-sequence variation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
The Joint United Nations Programme on HIV/AIDS. UNAIDS report on the global AIDS epidemic 2013 http://www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2013/gr2013/unaids_global_report_2013_en.pdf (2013)
Wyatt, R. & Sodroski, J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 (1998)
Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003)
Leonard, C. K. et al. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265, 10373–10382 (1990)
Cimbro, R. et al. Tyrosine sulfation in the second variable loop (V2) of HIV-1 gp120 stabilizes V2–V3 interaction and modulates neutralization sensitivity. Proc. Natl Acad. Sci. USA 111, 3152–3157 (2014)
Li, Y. et al. Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding, and intracellular transport. Proc. Natl Acad. Sci. USA 93, 9606–9611 (1996)
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998)
Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997)
Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997)
Julien, J. P. et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 (2013)
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 (2013)
Walker, L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011)
Huang, J. et al. Broad and potent neutralization of HIV-1 by a human antibody that recognizes an intersubunit site on the envelope glycoprotein. Nature http://dx.doi.org/10.1038/nature13601 (3 September 2014)
Sanders, R. W. et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76, 8875–8889 (2002)
Julien, J. P. et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl Acad. Sci. USA 110, 4351–4356 (2013)
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013)
Ringe, R. P. et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc. Natl Acad. Sci. USA 110, 18256–18261 (2013)
Munro, J. B. et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science http://dx.doi.org/10.1126/science.1254426 (2014)
Julien, J. P. et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9, e1003342 (2013)
Georgiev, I. S. et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756 (2013)
Mao, Y. et al. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nature Struct. Mol. Biol. 19, 893–899 (2012)
Pancera, M. et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl Acad. Sci. USA 107, 1166–1171 (2010)
Finzi, A. et al. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell 37, 656–667 (2010)
Buzon, V. et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 6, e1000880 (2010)
Caffrey, M. et al. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17, 4572–4584 (1998)
Nishikawa, K., Ooi, T., Saito, N. & Isogai, Y. Tertiary structure of proteins. 1. Representation and computation of conformations. J. Phys. Soc. Jpn. 32, 1331–1337 (1972)
Bartesaghi, A., Merk, A., Borgnia, M. J., Milne, J. L. & Subramaniam, S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nature Struct. Mol. Biol. 20, 1352–1357 (2013)
Liu, J., Bartesaghi, A., Borgnia, M. J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008)
White, T. A. et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6, e1001249 (2010)
Huang, C. C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 (2005)
McLellan, J. S. et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340, 1113–1117 (2013)
Yuan, W., Craig, S., Si, Z., Farzan, M. & Sodroski, J. CD4-induced T-20 binding to human immunodeficiency virus type 1 gp120 blocks interaction with the CXCR4 coreceptor. J. Virol. 78, 5448–5457 (2004)
Huang, C. C. et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317, 1930–1934 (2007)
Moore, J. P., McKeating, J. A., Weiss, R. A. & Sattentau, Q. J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250, 1139–1142 (1990)
Wilson, I. A., Skehel, J. J. & Wiley, D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289, 366–373 (1981)
Bullough, P. A., Hughson, F. M., Skehel, J. J. & Wiley, D. C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994)
McLellan, J. S., Yang, Y., Graham, B. S. & Kwong, P. D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 85, 7788–7796 (2011)
Weissenhorn, W., Carfi, A., Lee, K. H., Skehel, J. J. & Wiley, D. C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2, 605–616 (1998)
Lee, J. E. et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 (2008)
Colman, P. M. & Lawrence, M. C. The structural biology of type I viral membrane fusion. Nature Rev. Mol. Cell Biol. 4, 309–319 (2003)
Wyatt, R. et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393, 705–711 (1998)
Hraber, P. et al. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169 (2014)
Yang, X., Kurteva, S., Ren, X., Lee, S. & Sodroski, J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 79, 12132–12147 (2005)
Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002)
Klein, J. S. & Bjorkman, P. J. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6, e1000908 (2010)
Wu, X. et al. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80, 835–844 (2006)
McLellan, J. S. et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598 (2013)
Kanekiyo, M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013)
Liao, H.-X. et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013)
Doria-Rose, N. A. et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62 (2014)
McLellan, J. S. et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 (2011)
Majeed, S. et al. Enhancing protein crystallization through precipitant synergy. Structure 11, 1061–1070 (2003)
Kwong, P. D. & Liu, Y. Use of cryoprotectants in combination with immiscible oils for flash cooling macromolecular crystals. J. Appl. Cryst. 32, 102–105 (1999)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Adams, P. D. et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 (2004)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004)
Lin, C. W. & Ting, A. Y. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J. Am. Chem. Soc. 128, 4542–4543 (2006)
Zhou, Z. et al. Genetically encoded short peptide tags for orthogonal protein labeling by Sfp and AcpS phosphopantetheinyl transferases. ACS Chem. Biol. 2, 337–346 (2007)
Dave, R., Terry, D. S., Munro, J. B. & Blanchard, S. C. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009)
Aitken, C. E., Marshall, R. A. & Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008)
Richards, F. M. & Kundrot, C. E. Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins 3, 71–84 (1988)
Xiang, Z., Soto, C. S. & Honig, B. Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc. Natl Acad. Sci. USA 99, 7432–7437 (2002)
Xiang, Z. & Honig, B. Extending the accuracy limits of prediction for side-chain conformations. J. Mol. Biol. 311, 421–430 (2001)
Kirschner, K. N. et al. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 (2008)
Cornell, W. D. et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995)
Tomaras, G. D. et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82, 12449–12463 (2008)
Tomaras, G. D. et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J. Virol. 85, 11502–11519 (2011)
Li, M. et al. Human immunodeficiency virus type 1 env clones from acute and early subtype b infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 (2005)
Zhou, T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010)
Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007)
Scharf, L. et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 7, 785–795 (2014)
Calarese, D. A. et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071 (2003)
Xu, R. et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328, 357–360 (2010)
Ekiert, D. C. et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012)
Sui, J. et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature Struct. Mol. Biol. 16, 265–273 (2009)
Friesen, R. H. et al. A common solution to group 2 influenza virus neutralization. Proc. Natl Acad. Sci. USA 111, 445–450 (2014)
McLellan, J. S. et al. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nature Struct. Mol. Biol. 17, 248–250 (2010)
McLellan, J. S. et al. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J. Virol. 84, 12236–12244 (2010)
Mann, H. B. W. & Donald, R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60 (1947)
The PyMOL Molecular Graphics System. (DeLano Scientific, San Carlos, California, 2002)
Weis, W. I., Brunger, A. T., Skehel, J. J. & Wiley, D. C. Refinement of the influenza virus hemagglutinin by simulated annealing. J. Mol. Biol. 212, 737–761 (1990)
Chen, J., Skehel, J. J. & Wiley, D. C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl Acad. Sci. USA 96, 8967–8972 (1999)
Malashkevich, V. N. et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl Acad. Sci. USA 96, 2662–2667 (1999)
Lin, Y. P. et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc. Natl Acad. Sci. USA 109, 21474–21479 (2012)
Chen, L. et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326, 1123–1127 (2009)
Rini, J. M. et al. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc. Natl Acad. Sci. USA 90, 6325–6329 (1993)
Stanfield, R. et al. Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs. Structure 7, 131–142 (1999)
Tugarinov, V., Zvi, A., Levy, R. & Anglister, J. A cis proline turn linking two β-hairpin strands in the solution structure of an antibody-bound HIV-1IIIB V3 peptide. Nature Struct. Biol. 6, 331–335 (1999)
Ofek, G. et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78, 10724–10737 (2004)
Cardoso, R. M. et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173 (2005)
Luftig, M. A. et al. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nature Struct. Mol. Biol. 13, 740–747 (2006)
Cardoso, R. M. et al. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 365, 1533–1544 (2007)
Grigoryan, G. & Keating, A. E. Structural specificity in coiled-coil interactions. Curr. Opin. Struct. Biol. 18, 477–483 (2008)
Helseth, E., Olshevsky, U., Furman, C. & Sodroski, J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65, 2119–2123 (1991)
Thali, M., Furman, C., Helseth, E., Repke, H. & Sodroski, J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J. Virol. 66, 5516–5524 (1992)
Cao, J. et al. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67, 2747–2755 (1993)
Leavitt, M., Park, E. J., Sidorov, I. A., Dimitrov, D. S. & Quinnan, G. V., Jr Concordant modulation of neutralization resistance and high infectivity of the primary human immunodeficiency virus type 1 MN strain and definition of a potential gp41 binding site in gp120. J. Virol. 77, 560–570 (2003)
Yang, X., Mahony, E., Holm, G. H., Kassa, A. & Sodroski, J. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313, 117–125 (2003)
Sen, J., Jacobs, A. & Caffrey, M. Role of the HIV gp120 conserved domain 5 in processing and viral entry. Biochemistry 47, 7788–7795 (2008)
Wang, J., Sen, J., Rong, L. & Caffrey, M. Role of the HIV gp120 conserved domain 1 in processing and viral entry. J. Biol. Chem. 283, 32644–32649 (2008)
Lawless, M. K. et al. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry 35, 13697–13708 (1996)
Chen, C. H., Matthews, T. J., McDanal, C. B., Bolognesi, D. P. & Greenberg, M. L. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69, 3771–3777 (1995)
Root, M. J., Kay, M. S. & Kim, P. S. Protein design of an HIV-1 entry inhibitor. Science 291, 884–888 (2001)
Gustchina, E. et al. Structural basis of HIV-1 neutralization by affinity matured Fabs directed against the internal trimeric coiled-coil of gp41. PLoS Pathog. 6, e1001182 (2010)
Sabin, C. et al. Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41. PLoS Pathog. 6, e1001195 (2010)
Blattner, C. et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40, 669–680 (2014)
Yasmeen, A. et al. Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology 11, 41 (2014)
Falkowska, E. et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40, 657–668 (2014)
Thali, M. et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67, 3978–3988 (1993)
Guttman, M. et al. CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22, 974–984 (2014)
Scheid, J. F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011)
Walker, L. M. et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009)
Stanfield, R. L. & Wilson, I. A. Structural studies of human HIV-1 V3 antibodies. Hum. Antibodies 14, 73–80 (2005)
Rizzuto, C. D. et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280, 1949–1953 (1998)
Guan, Y. et al. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl Acad. Sci. USA 110, E69–E78 (2013)
Gorny, M. K., VanCott, T. C., Williams, C., Revesz, K. & Zolla-Pazner, S. Effects of oligomerization on the epitopes of the human immunodeficiency virus type 1 envelope glycoproteins. Virology 267, 220–228 (2000)
Yuan, W. et al. Oligomer-specific conformations of the human immunodeficiency virus (HIV-1) gp41 envelope glycoprotein ectodomain recognized by human monoclonal antibodies. AIDS Res. Hum. Retroviruses 25, 319–328 (2009)
Moore, P. L. et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80, 2515–2528 (2006)
Frey, G. et al. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nature Struct. Mol. Biol. 17, 1486–1491 (2010)
Miller, M. D. et al. A human monoclonal antibody neutralizes diverse HIV-1 isolates by binding a critical gp41 epitope. Proc. Natl Acad. Sci. USA 102, 14759–14764 (2005)
Chen, J. et al. Mechanism of HIV-1 neutralization by antibodies targeting a membrane-proximal region of gp41. J. Virol. 88, 1249–1258 (2014)
Frey, G. et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl Acad. Sci. USA 105, 3739–3744 (2008)
Nicely, N. I. et al. Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region. Nature Struct. Mol. Biol. 17, 1492–1494 (2010)
Huang, J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012)
Chakrabarti, B. K. et al. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res. Hum. Retroviruses 27, 877–887 (2011)
Ruprecht, C. R. et al. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J. Exp. Med. 208, 439–454 (2011)
Acknowledgements
We thank J. Binley for the JR-FL plasmid used in smFRET; D. Burton and W. Koff for PGT122; J. Chrzas and SER-CAT staff for assistance with data collection; B. Graham for discussions on RSV; R. Sanders for furin plasmid; R. Schwartz and the Vaccine Production Program for antibody VRC01; J. Sodroski for information on C34-Ig; I. Wilson for discussions on pre-fusion Env structure; Z. Yang for assistance with cloning; G. Nabel, G. Scott and members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions and comments on the manuscript; and the WCMC/AMC/TSRI HIVRAD team for their contributions to the design and validation of near-native mimicry for soluble BG505 SOSIP.664 trimers. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panel. The authors acknowledge the contributions of the Center for HIV/AIDS Vaccine Discovery (CHAVI) Clinical Core Team for recruiting study participants and carrying out all aspects of the CHAVI001 and CHAVI008 protocols at Chapel Hill, North Carolina (J. Eron); Blantyre, Malawi (J. Kumwenda, T. Taha); Lilongwe, Malawi (I. Hoffman, G. Kaminga); Johannesburg, South Africa (H. Rees); Durban, South Africa (S. Abdool Karim); Moshi, Tanzania (S. Noel, S. Kapiga, J. Crump); and London, UK (S. Fidler). Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH); and by grants from the Division of AIDS, NIAID, NIH (R21-AI100696, CHAVI-AI0678501, and CHAVI-Immunogen Discovery-AI100645), from the National Institutes of General Medical Sciences (PO1-GM56550 and RO1-GM098859), from the Irvington Fellows Program of the Cancer Research Program, and from the International AIDS Vaccine Initiative’s (IAVI’s) Neutralizing Antibody Consortium. IAVI's work is made possible by support from many donors including: the Bill & Melinda Gates Foundation; the Ministry of Foreign Affairs of Denmark; Irish Aid; the Ministry of Finance of Japan; the Ministry of Foreign Affairs of the Netherlands; the Norwegian Agency for Development Cooperation (NORAD); the UK Department for International Development (DFID): and the United States Agency for International Development (USAID). The full list of IAVI donors is available at http://www.iavi.org. Use of sector 22 (Southeast Region Collaborative Access Team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Author information
Authors and Affiliations
Contributions
M.P., T.Z., A.D. and P.D.K. determined the trimer structure, with M.P. heading structure determination; and with T.Z. assisting with solution and refinement, A.D. with protein production, and P.D.K. with data collection. I.S.G. and C.S. performed bioinformatics analysis, J.G. performed conformational analysis, P.A., G.-Y.C., G.O., G.B.E.S.-J. and J.S. performed antigenic and mechanistic analyses, R.T.B., M.G.J., M.K.L., N.T., Y.Y., B.Z., M.S.C., B.F.H., J.R.M. and L.M. performed cohort analysis, J.B.M., S.C.B. and W.M. performed smFRET analysis, and J.H. and M.C. provided antibody 35O22. M.P. and P.D.K. wrote the paper, on which all principal investigators commented.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
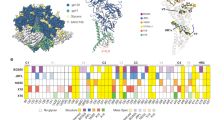
Extended Data Figure 1 Antibody-mediated crystallization and antibody-induced conformation.
a, Atomic-level structures for HIV-1 Env regions determined in complex with HIV-1-neutralizing antibodies. Neutralizing antibodies generally recognize the pre-fusion conformation of HIV-1 Env. Structures highlighted here display a cumulative sum total of pre-fusion HIV-1 Env structural information. Env residues are numbered according to standard HX numbering (from PDBs). One structure, for antibody D5 (blue), is in the post-fusion gp41 conformation, and is not included in the sum total. Regions of other structures (purple) did not define sequence register, and were also not included in the sum total. References listed in this figure are cited elsewhere in the manuscript, except for Rini et al. (1993)87, Stanfield et al. (1999)88,89, Ofek et al. (2004)90, Cardoso et al. (2005)91, Luftig et al. (2006)92, Cardoso et al. (2007)93. b, Antibody-induced conformation of HIV-1 Env in the context of infectious JR-FL virions as assessed by smFRET. HIV-1JR-FL gp160 was labelled with fluorescent dyes in variable regions, V1 and V4, at positions that did not interfere with Env function (see Methods), and virus was surface-immobilized for imaging via total internal reflection fluorescence microscopy18. smFRET trajectories were compiled into histograms for the HIV-1JR-FL Env trimer, either unliganded or after pre-incubation for 30 min with 0.1 mg ml−1 PGT122, 35O22, or both PGT122 and 35O22 before imaging. Resultant Env conformational landscapes could be deconvoluted into three gaussian distributions: a low-FRET population that predominated for the pre-fusion mature unliganded state, and intermediate- and high-FRET populations, which predominated in the presence of CD4 receptor and CD4-induced antibody18. smFRET trajectories are shown for the unliganded HIV-1JR-FL Env trimer as well as in the presence of PGT122, 35O22, and both PGT122 and 35O22. The concordance between conformational ensembles indicates unliganded and PGT122+35O22-bound conformation to be similar (Spearman correlation coefficient of 0.988). Interestingly, the presence of just one of the antibodies (PGT122) appeared to reduce the high FRET population, an effect not observed in the presence of both antibodies; this suggests that the antibody-induced stability of a particular state is not solely additive, and that antibodies can both induce a particular conformational state as well as alter the transition dynamics from that state.
Extended Data Figure 2 HIV-1 subunit interactions: principle component analysis and interface contacts.
a, Minimum-bounding box, generated by principle component analysis, encasing 90% of the HIV-1 Env gp120–gp41 protomer. Each gp120–gp41 blade forms a rectangle of height of ∼100 Å, width of ∼65 Å, and thickness of ∼35 Å. Subunits are displayed in ribbon representation with gp41 coloured rainbow and gp120 coloured and labelled red. As previously visualized10,11, the membrane-distal portion of the rectangle is made up of the gp120-outer and -inner domains, with the central 7-stranded β-sandwich of the inner domain occupying the trimer-distal, membrane-proximal portion of gp120. We have now resolved the rest of the spike: the membrane-proximal portion of the rectangle is made up of gp41, with the membrane-distal portion of gp41 closest to the molecular threefold axis occupied by helix α7 (which corresponds in register to the C-terminal portion of the post-fusion HR1 helix of gp41), and the rest of gp41 folding around N and C termini strands of gp120, which extend over 20 Å towards the viral membrane. Of the four helices, α6 kinks at residue 537gp41 and α9 kinks at residue 637gp41; backbone H-bonding is less ideal at residues 663gp41 and 664gp41. b, Different views of trimeric protomer association. The protomer association at the membrane-distal trimer apex occurs through the corners of the minimum-bounding box, whereas the association at the membrane-proximal region occurs with substantial interpenetration of the minimum-bounding box; these interaction differences and the protruding nature of the gp120 outer domain result in the overall mushroom shape of the trimer. c, gp120–gp41 interface. Ribbon representation of gp120 (red) and gp41 (rainbow from blue N terminus to orange C terminus), with gp120 residues that interact with gp41 shown in surface representation and gp41 residues that interact with gp120 shown in semi-transparent surface. A complete list of subunit interactions is provided in Supplementary Table 1. Membrane-proximal interactions are further stabilized by hydrophobic interactions, which gp41 makes with the N and C termini of gp120, such as between Trp 35gp120 and Pro609gp41 and between Trp 610gp41 and Pro498gp120. d, Wheel diagram representation of α7 coiled-coil in the pre-fusion mature closed conformation of gp41 as generated by DrawCoil 1.0: http://www.grigoryanlab.org/drawcoil/(ref. 94). e, gp41–trimer interfaces as viewed from the viral membrane in ribbon and surface representation (90° rotation from Fig. 2c). f, BG505 SOSIP.664 sequence with residues identified by mutagenesis95,96,97,98,99,100,101 to be important for gp120/gp41 association underlined. Residues that were found to interact between gp120 and gp41 by examination of the crystal structure are indicated in red (intra-protomer interactions) and in brown (inter-protomer interactions). Sites of N-linked glycosylation are shown in green; glycan N88 is shown in red because it is part of the gp120/gp41 interactions; no density was observed for potential N-linked glycans at residues 185, 398, 406, 411, 462 and 625. Residues that were disordered in the crystal structure are grey. SOS (A501C/T605C) and IP (I559P) mutations are labelled in bold and italics. Dots indicate residues not present in the BG505 sequence.
Extended Data Figure 3 Modelling of gp41: pre-fusion α6-to-α7 density, HIV-1–SIV post-fusion chimaera, and liganded interactions.
a, Modelling of gp41 residues 548–568. At low contour, suggestive density is observed that might correspond to the connection between α6 and α7 helices. This density appeared to be crystal dependent and might be related to inherent flexibility, functional rearrangements, asymmetry between protomers, or combinations of these factors. To investigate the degree to which a model for this region might be defined, we built and refined two different models for this region: electron density (blue) shown for 2Fo − Fc density at 1σ contour; gp41 (rainbow colour from blue to orange) shown in ribbon representation with side chains; gp120 (red) shown in ribbon representation. The location of the I559P mutation is indicated. b, The two models from panel a are superimposed and shown in perpendicular orientations. c, HIV-1–SIV post-fusion chimaera. Sequences of HIV-1 gp41 from pre-fusion structure (BG505 strain, PDB ID 4TVP), post-fusion structure (HIVpost, PDB ID 2X7R24) and SIV gp41 post-fusion structure (SIVpost, PDB ID 2EZO25) are aligned with secondary structure indicated. Residues that were used to make the post-fusion HIV-1–SIV chimaera used in Fig. 3 are highlighted in red. d, Binding residues of representative fusion-intermediate entry inhibitors or antibodies mapped onto the structure of pre-fusion HIV-1 Env spike102,103,104. Top, ribbon representation of pre-fusion envelope protomer A (gp120 in red and gp41 in blue) at two orientations, with the binding residues of the fusion-intermediate inhibitors 5-helix92 and T20102,103 and of monoclonal antibody D592 shown in orange, green and yellow, respectively. Bottom, surface representation of the pre-fusion envelope trimer, with inhibitor and antibody binding residues mapped onto the surfaces of all protomers (A, B, C). gp120 is coloured grey and gp41 is coloured in shades of blue, depending on protomer. Binding residues of fusion-intermediate inhibitors 5-helix, T20 and monoclonal antibody D5 are shown in same colour shades as in the top panels. e, 5-helix, T20 and D5 Fab (all coloured magenta and grey) docked onto a model of fusion-intermediate gp41 (coloured as in d). f, A previously defined binding pocket on post-fusion gp41 is recognized by pre-fusion gp41 tryptophan-clasp residues Trp 628 and Trp 631. Shown is a surface representation of gp41 5-helix protein104 (left, with N-heptad repeat (NHR) helices coloured in shades of green and C-heptad repeat (CHR) helices coloured in shades of orange). The footprint of gp41 tryptophan-clasp residues Trp 628 and Trp 631 is shown in magenta (middle) and that of a representative NHR-specific neutralizing antibody, D5, in yellow92,105,106 (right).
Extended Data Figure 4 Conformational changes between pre-fusion mature closed state and CD4-bound state of gp120.
a, Overall structure and sequence comparison. gp120 is shown in ribbon representation in pre-fusion mature closed (red) and CD4-bound (yellow, PDB ID 3JWD22) conformation. V1V2 (PDB ID 3U2S51) and V3 (PDB ID 2B4C30) have been modelled onto the CD4-bound conformation. Secondary structure is defined for pre-fusion and CD4-bound conformation on the BG505 sequence, with cylinders representing α-helix and arrows β-strands. Disordered residues are indicated by X. Residues that move more than 3 Å between the mature closed and the CD4-bound gp120 conformations are highlighted by grey shadows. Sites of N-linked glycosylation are shown in green. b, Details of conformational changes between the mature closed (red) and the CD4-bound conformations (yellow) of gp120 (shown in ribbon). Regions highlighted cover layer 1 with changes at α0 (we note that density in this region is not well defined), layer 2 with changes at α1 and β20-21 rearrangements. All atoms r.m.s.d. are as follows: residues 54gp120–74gp120, r.m.s.d. = 4.759 Å; residues 98gp120–117gp120, r.m.s.d. = 0.497 Å; 424gp120–436gp120, r.m.s.d. = 3.196 Å.
Extended Data Figure 5 Antigenic profiles of HIV-1 envelope conformational states.
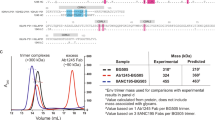
a, Qualitative recognition of HIV-1 envelope by diverse antibodies is shown for five conformational states. Green bars indicate reported recognition and red bars no recognition; absence of a bar indicates that recognition is undefined. The compiled data are from cited references and experiments described in this figure. Note references 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127 are cited here. b, Octet Biosensorgrams of BG505 SOSIP.664 (left) and BG505 gp120 (right) binding to human monoclonal IgGs. The dotted line indicates the beginning of the dissociation phase and the maximal specific binding after 300 s reported in the table (−, <0.05 response units (RU); +, 0.05 RU to 0.25 RU; ++, 0.25 RU to 0.5 RU; and +++, >0.5 RU). BG505gp120 did not contain the T332N mutation (no glycan at that position). Both proteins were made in GnTi−/−. We note that antigenicity of the BG505 SOSIP.664 and BG505 gp120 protein varied depending on the assay done. Thus, using surface plasmon resonance (SPR), no CD4i antibody binding was detected while some binding could be observed using biolayer interferometry. Although PG9 bound BG505 gp120 in ELISA16, it did not bind in biolayer interferometry format. We observed 447-52D binding, while it was not observed in previously published ELISA16. c, SPR binding affinities of 35O22, PGT151 and PGT145 to BG505 SOSIP.664 and influence of sCD4. d, Estimation of binding stoichiometry for 35O22, PGT151, and PGT145 to trimeric BG505 SOSIP.664 by SPR and comparison to published data13,107,108. e, Effect of sCD4 and sCD4/17b on binding of antibodies 35O22 and PGT151 to BG505 SOSIP.664 by SPR. The structure of a pre-fusion mature closed state of HIV-1 provides a critical addition to the pantheon of HIV-1 Env structures with atomic-level detail. Moreover, antibodies 35O22 and PGT151, which bind specifically to the trimeric pre-fusion conformation of gp41, provide new tools by which to assess the conformational state of gp4113,107,109. The binding of antibodies 35O22 and PGT151 to BG505 SOSIP.664 trimer was tested in the presence of the CD4 receptor and the 17b antibody110 (a co-receptor surrogate which recognizes a bridging sheet epitope that overlaps the site of co-receptor recognition). In the case of antibody 35O22, CD4 binding to the BG505 SOSIP.664 trimer affected the kinetics, affinity and stoichiometry of binding. 35O22 bound to BG505 SOSIP.664 with an 8.4-fold reduced affinity, primarily contributed by an increased rate of dissociation. The overall binding level (Rmax) normalized to the average level of trimer captured (see also panel d) was lower, suggesting substoichiometric binding. Capturing the trimer on a CD4–Ig surface reduced normalized Rmax for PGT151 compared to the 2G12 capture format, suggesting reduced stoichiometry for PGT151 binding to trimer pre-bound with CD4, although kinetics and affinity of interaction were similar. A BG505 SOSIP.664 trimer + sCD4 complex captured onto a 17b surface-bound 35O22 but showed no detectable binding to PGT151.
Extended Data Figure 6 N-linked glycan occlusion of type I fusion machines.
The pre-fusion mature closed conformation of HIV-1 Env evades the humoral immune response with a fully assembled glycan shield. Here we calculate and display the solvent-accessible surface of glycan and protein for HIV-1 Env, HIV241 (which contains an added glycan at position N241), influenza virus haemagglutinin and RSV fusion glycoprotein. Calculations of the percentage coverage of the protein surface were determined for trimeric type I fusion machines based on two probe sizes of 1.4 Å (solvent radius) and 10.0 Å (the estimated steric footprint of an antibody combining region). Surface area calculations were carried out according to Kong et al.79, and images were generated using Grasp v1.380. All models were refined using Amber with the GLYCAM force field (see Methods for details). The PDB IDs associated with the glycosylated models are: 4TVP (HIV-1), 2YP785 (Flu) and 4JHW31 (RSV). The strains associated with the PDB IDs are: BG505.SOSIP.664 (HIV-1), H3N2 A/Hong Kong/4443/2005 (Flu) and A/A2/61 (RSV). The solvent-accessible protein surface is shown in red, and N-linked glycans are shown in green. a, Estimated Man9 glycan coverage. b, Estimated Man5 glycan coverage. c, Visualization of Man9 N-linked glycan coverage for two probe radii. d, Visualization of Man5 N-linked glycan coverage for two probe radii.
Extended Data Figure 7 Glycan shield and sequence variability for HIV-1 pre-fusion mature closed and CD4-bound conformations.
Many conformations of HIV-1 Env divert the immune response. Thus for example, shed gp120 and post-fusion gp41 represent dominant viral antigens; however, these forms of Env are not functional, and antibodies that only target them are not neutralizing. Functional conformations, however, may be significantly shielded from the neutralizing antibody. The CD4-bound conformation of HIV-1 Env, for example, is only functionally present when the viral and target-cell membranes are in close proximity, and the exposed co-receptor binding site (including V3- and CD4-induced epitopes) is spatially occluded from neutralizing antibody. Here we provide models for the pre-fusion closed state versus the CD4-bound conformation, which display the fully assembled glycan shield and surface Env variability. Env N-linked glycans are depicted in light green (conserved; greater than 90% conservation) or dark green (variable; less than 90% conservation) on the mature closed Env structure and modelled CD4-bound conformation. Env sequence variability is shown from white to purple (conserved to variable). A conserved glycan at residue 241gp120 not present in the BG505 sequence is shown in yellow-green. As can be seen, the pre-fusion closed state has few glycan-free surfaces, whereas the CD4-bound state exposes substantial glycan-free conserved surface.
Extended Data Figure 8 Prevalence of neutralizing responses identified serologically from cohorts from 2–3 years and 5+ years post infection.
a, Serum neutralization on 21-strain virus panel. ID50s (reciprocal dilution at which 50% of the virus is neutralized) are shown for serum (rows) titrated against HIV-1 viral strains (columns). b, For each serum, the predicted neutralization prevalence for each of 12 antibody specificities is shown based on neutralization of 21 diverse HIV-1 strains. Values of at least 0.2 were considered positive and counted toward the overall cohort prevalence percentages in Fig. 6c. c, Prevalence of antibody specificities onto the HIV-1 Env, coloured as indicated in the bar graph. d, The antibody specificities for high serum prevalence in the 5+ years cohort are depicted by Fabs of representative antibodies binding the BG505 SOSIP.664 Env trimer, shown in grey ribbon representation, with glycans as green sticks. Note that while prevalence between the two cohorts showed good correspondence, there were notable differences, for example, between PGT151 at 2–3 years and 5+ years in this study as well as between the cohorts analysed here and in ref. 13.
Extended Data Figure 9 Antibodies 35O22 and PGT122: interface with HIV-1 Env and comparison of bound and unbound Fab conformations.
Despite the substantial immune evasion protecting the mature unliganded state from humoral recognition, after several years of infection, the human immune system does generate broadly neutralizing antibodies. 35O22 and PGT122 are two of these antibodies, which neutralize 62% and 65% of HIV-1 isolates at a median IC50(half maximal inhibitory concentration) of 0.033 and 0.05 µg ml−1, respectively13,12. Here we provide additional details on 35O22 and PGT122 recognition. a, 35O22 Fab is shown in ribbon representation (purple (heavy chain) and white (light chain)). The gp120 subunit is shown in red, the gp41 subunit in rainbow (from blue N terminus to orange C terminus), and glycans in green sticks. Complementarity determining regions (CDRs) are labelled, and interactive HIV-1 Env residues highlighted in semi-transparent surface representation. At the membrane-distal surface of 35O22, an extended framework 3 region (FW3) of the heavy chain (resulting from an insertion of 8 residues) interacts with strand β1 of the 7-stranded inner domain sandwich of gp120. The heavy chain-CDRs form extensive contacts with the N-linked glycan extending from residue 88gp120. In addition to glycan contacts, the CDR H3 of 35O22 interacts with the α9 helix of gp41. Helix α9 interactions are also made by the FW3 of the light chain (a complete list of contacts is provided in Supplementary Table 3). Overall, 35O22 buries 1,105 Å2 solvent surface on gp120 (including 793 Å2 with the Asn 88gp120 glycan) and 594 Å2 solvent surface on gp41 (including 127 Å2 with the Asn 618gp41 glycan). Despite residue 625gp41 being part of the glycan sequon ‘NMT’, no glycan is observed; indeed, the side-chain amide of residue 625gp41 hydrogen bonds with the side-chain oxygen of Tyr 32 in the 35O22 heavy chain, and the presence of an N-linked glycan at residue 625gp41 is difficult to reconcile with 35O22 recognition. b, Same colours as a, with 35O22 Fab shown in surface representation. c, Same colours as a, with 2Fo − Fc at 1σ contour (blue density) shown around glycan 88 of gp120. Antibody 35O22 employs a novel mechanism of glycan-protein recognition, combining a protruding FW3 with CDR H1, H2 and H3 to form a ‘bowl’ that holds glycan. FW3 and CDR H3 provide the top edges of the bowl and interact with the protein surface of gp120, whereas CDR H1 and H2 are recessed and hold/recognize glycan. This structural mechanism of recognition contrasts with the extended CDR H3-draping glycan observed with other antibodies that penetrate the glycan shield such as PG951 and PGT12878. d, Unbound and HIV-1 Env-bound 35O22 Fabs were superimposed, and ribbon representations and r.m.s.d.s are displayed. Unbound 35O22 Fab is coloured cyan (heavy chain) and green (light chain), and bound 35O22 Fab is coloured deep purple (heavy chain) and white (light chain). Regions that showed conformational changes are highlighted with black dotted lines. We note that in the 35O22-bound conformation, density is poor and/or sparse for the Fc portion of the Fab. e, PGT122 interface details. Ribbon representation of PGT122 Fab in blue (heavy chain) and light blue (light chain) interacting with one gp120 subunit, shown in red with glycans in green sticks. CDRs are labelled, and interactive HIV-1 Env residues highlighted in surface representation. Primary contacts between antibody PGT122 and N-linked glycan involve N137 and N332, with minor contact with N156. Although portions of glycan N301 can be observed in the electron density, no direct contacts with PGT122 are observed; a complete list of contacts between PGT122 and BG505 SOSIP.664 is provided in Supplementary Table 4. f, Same colours as e, with PGT122 Fab shown in surface representation. g, Same colours as e, with 2Fo − Fc at 1σ contour (grey density) shown around glycan 332 of gp120. h, Comparison of bound and unbound PGT122 Fab conformations. Unbound and HIV-1 Env-bound Fabs were superimposed, and ribbon representations and r.m.s.d.s are displayed. Unbound PGT122 Fab is coloured cyan, and bound PGT122 Fab blue (heavy chain) and light blue (light chain). Regions which showed conformational changes are highlighted with black dotted lines.
Extended Data Figure 10 Structural implementation of HIV-1 molecular trickery.
The pre-fusion HIV-1 Env trimer (left) is displayed with evasion mechanisms and their structural implementation (right). The gp120 subunit is shown in red, the gp41 subunit in rainbow (from blue N terminus to orange C terminus), and crystallographically defined glycans in green. One protomer is shown with Cα trace and glycans in stick representation; a second protomer is shown in ribbon representation with secondary structure elements labelled; and the third protomer is shown in light grey surface. The MPER region for each protomer is shown as a stylized helix associated with the viral membrane. The location of secondary structural elements, termini, and residues called in the text has been labelled (red font for gp120 and black font for gp41).
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4. (PDF 674 kb)
Rights and permissions
About this article
Cite this article
Pancera, M., Zhou, T., Druz, A. et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014). https://doi.org/10.1038/nature13808
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13808
This article is cited by
-
Conformational antigenic heterogeneity as a cause of the persistent fraction in HIV-1 neutralization
Retrovirology (2023)
-
HIV-1 Env trimers asymmetrically engage CD4 receptors in membranes
Nature (2023)
-
Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs
Nature Communications (2023)
-
Strategies for HIV-1 vaccines that induce broadly neutralizing antibodies
Nature Reviews Immunology (2023)
-
Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization
Nature Structural & Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.