Abstract
The semi-conservative centrosome duplication in cycling cells gives rise to a centrosome composed of a mother and a newly formed daughter centriole1. Both centrioles are regarded as equivalent in their ability to form new centrioles and their symmetric duplication is crucial for cell division homeostasis2,3,4. Multiciliated cells do not use the archetypal duplication program and instead form more than a hundred centrioles that are required for the growth of motile cilia and the efficient propelling of physiological fluids5. The majority of these new centrioles are thought to appear de novo, that is, independently from the centrosome, around electron-dense structures called deuterosomes6,7,8. Their origin remains unknown. Using live imaging combined with correlative super-resolution light and electron microscopy, we show that all new centrioles derive from the pre-existing progenitor cell centrosome through multiple rounds of procentriole seeding. Moreover, we establish that only the daughter centrosomal centriole contributes to deuterosome formation, and thus to over ninety per cent of the final centriole population. This unexpected centriolar asymmetry grants new perspectives when studying cilia-related diseases5,9 and pathological centriole amplification observed in cycling cells and associated with microcephaly and cancer2,3,4,10.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Vorobjev, I. A. & Chentsov Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93, 938–949 (1982)
Lingle, W. L. et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl Acad. Sci. USA 99, 1978–1983 (2002)
Ganem, N. J., Godinho, S. A. & Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009)
Marthiens, V. et al. Centrosome amplification causes microcephaly. Nature Cell Biol. 15, 731–740 (2013)
Sharma, N., Berbari, N. F. & Yoder, B. K. Ciliary dysfunction in developmental abnormalities and diseases. Curr. Top. Dev. Biol. 85, 371–427 (2008)
Sorokin, S. P. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3, 207–230 (1968)
Zhao, H. et al. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nature Cell Biol. 15, 1434–1444 (2013)
Klos Dehring, D. A. et al. Deuterosome-mediated centriole biogenesis. Dev. Cell 27, 103–112 (2013)
Wallmeier, J. et al. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nature Genet. 46, 646–651 (2014)
Godinho, S. A. et al. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510, 167–171 (2014)
Anderson, R. G. & Brenner, R. M. The formation of basal bodies (centrioles) in the rhesus monkey oviduct. J. Cell Biol. 50, 10–34 (1971)
Dirksen, E. R. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J. Cell Biol. 51, 286–302 (1971)
Loots, G. P. & Nel, P. P. Early stages of ciliogenesis in the respiratory epithelium of the nasal cavity of rabbit embryos. Cell Tissue Res. 255, 589–594 (1989)
Higginbotham, H., Bielas, S., Tanaka, T. & Gleeson, J. G. Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res. 13, 155–164 (2004)
Piel, M., Meyer, P., Khodjakov, A., Rieder, C. L. & Bornens, M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells 7. J. Cell Biol. 149, 317–330 (2000)
Wang, X. et al. Asymmetric centrosome inheritance maintains neural progenitors in neocortex. Nature 461, 947–955 (2009)
Anderson, C. T. & Stearns, T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 19, 1498–1502 (2009)
Kitagawa, D. et al. Structural basis of the 9-fold symmetry of centrioles. Cell 144, 364–375 (2011)
Dzhindzhev, N. S. et al. Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718 (2010)
Habedanck, R., Stierhof, Y.-D., Wilkinson, C. J. & Nigg, E. A. The Polo kinase Plk4 functions in centriole duplication. Nature Cell Biol. 7, 1140–1146 (2005)
Singla, V. & Reiter, J. F. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science 313, 629–633 (2006)
Robbins, E., Jentzsch, G. & Micali, A. The centriole cycle in synchronized HeLa cells. J. Cell Biol. 36, 329–339 (1968)
Azimzadeh, J. et al. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185, 101–114 (2009)
Wolff, A. et al. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59, 425–432 (1992)
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012)
Hoh, R. A., Stowe, T. R., Turk, E. & Stearns, T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE 7, e52166 (2012)
Stubbs, J. L., Vladar, E. K., Axelrod, J. D. & Kintner, C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nature Cell Biol. 14, 140–147 (2012)
Tan, F. E. et al. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development 140, 4277–4286 (2013)
Marcet, B. et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nature Cell Biol. 13, 693–799 (2011)
Ma, L., Quigley, I., Omran, H. & Kintner, C. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 28, 1461–1471 (2014)
Marszalek, J. R., Ruiz-Lozano, P., Roberts, E., Chien, K. R. & Goldstein, L. S. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl Acad. Sci. USA 96, 5043–5048 (1999)
Mirzadeh, Z., Doetsch, F., Sawamoto, K., Wichterle, H. & Alvarez-Buylla, A. The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 39, 1938 (2010)
Lin, Y. N. et al. CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell Biol. 202, 211–219 (2013)
Graser, S. et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330 (2007)
Hung, L. Y., Tang, C. J. & Tang, T. K. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the γ-tubulin complex. Mol. Cell. Biol. 20, 7813–7825 (2000)
Bosch Grau, M. et al. Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J. Cell Biol. 202, 441–451 (2013)
Delgehyr, N., Sillibourne, J. & Bornens, M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118, 1565–1575 (2005)
Kleylein-Sohn, J. et al. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190–202 (2007)
Acknowledgements
We thank P. Rostaing of the IBENS Electron microscopy facility for making the correlative 3D-SIM and electron microscopy experiments possible. We also thank the Imaging Facility of IBENS, which is supported by grants from Fédération pour la Recherche sur le Cerveau, Région Ile de France DIM NeRF 2009 and 2011 and France-BioImaging. We wish to thank L. Sengmanivong from the Nikon Imaging Centre at Institut Curie-CNRS. We are grateful to L. Goldstein for providing us with the Kif3a mutant mice. We thank C. Janke for the GT335 antibody, E. A. Nigg for the Cep164 and Plk4 antibodies, T. K. Tang for Cep120 and Cpap antibodies, X. Yan and X. Zhu for Deup1 and Cep152 antibodies, J. Azimzadeh for the Poc5 antibody and X. Morin for his gift of the pCAAGS-Cen2–GFP-mCherry plasmid. We also thank M. Bornens, B. Guirao, J. B. Boulé, C. Janke and J.-F. Brunet for their comments on the manuscript and all the members of the Spassky laboratory for discussions. This study was supported by the CNRS, the ENS, INSERM, an FPGG grant, ANR award ANR-12-BSV4-0006 CILIASTEM, ARC award PJA-20131200184, La ligue contre le cancer-comité de Paris RS14/75-88, the ‘Investissements d’Avenir’ program of the French Government and implemented by the ANR (referenced ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001-02 PSL* Research University), a start-up grant from the City of Paris and a start-up grant from the FRM. A.A.J. and A.-I.L. received fellowships from the French Ministry of Higher Education and Research.
Author information
Authors and Affiliations
Contributions
A.A.J. performed and analysed experiments; A.-I.L. performed videomicroscopy experiments; N.D. performed and analysed cilia mutant experiments; M.F. contributed to in vitro experiments; N.S. designed the study and supervised the project; A.M. initiated the study, designed, performed and analysed experiments, and supervised the project; A.M., N.S. and A.A.J. wrote the manuscript. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
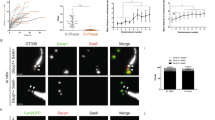
Extended Data Figure 1 Centriole amplification in ependymal progenitor cells in vitro and in vivo.
a, TEM showing the apical surface of a differentiating ependymal progenitor in vitro (DIV 3). Procentrioles (arrowheads) are seen growing from deuterosomes (D; ‘deuterosome’ pathway) or from a centriolar structure (C; ‘centriolar’ pathway). b, Ki67, Sas-6 and GT335 immunoreactivity during centrosome duplication in Cen2–GFP+ cycling ependymal progenitors in vitro (DIV −2). c–e, FoxJ1, Sas-6, GT335 and Cep164 immunoreactivity in Cen2–GFP+ differentiating ependymal progenitors in vitro (DIV 2 to DIV 5) in order of appearance (see h). f, In vivo immunostainings of Sas-6 and FoxJ1 in P3 to P5 Cen2–GFP+ ventricular walls showing that the same stages of amplification exist in vivo as in vitro (see also h). Boxes in b–f indicate magnified structures. White arrowheads indicate the centrosomal centrioles at the halo stage. g, Analysis in vitro (DIV 3) and in vivo (P3–P5 lateral ventricular walls) of Cen2–GFP fluorescence showing an increase in the intensity and area of Cen2–GFP ‘flower’ fluorescence compared to Cen2–GFP ‘halos’ (in vitro: n = 48 halos; n = 47 flowers; in vivo: n = 37 halos; n = 27 flowers); error bars represent mean ± s.d.; P values derived from two-tailed Mann–Whitney U-tests, ***P < 0.0001. h, Left: percentages of differentiating cells in vitro according to DIV (n = 1,244 cells from 3 experiments per day). Right: percentages of differentiating cells in vivo in the region of the lateral brain ventricle indicated by the red square in Fig. 1a according to post-natal day (P) (n = 4,025 cells from 2 to 3 animals per age). Scale bars: 1 µm.
Extended Data Figure 2 Dynamics of centriole amplification analysed live in vitro; centrosomes of cycling ependymal progenitors in vitro; centrosome Sas-6 asymmetry in vivo.
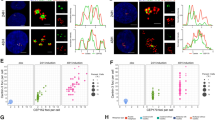
a, From Supplementary Video 2. Dynamics of centriole amplification in a differentiating Cen2–GFP+ ependymal progenitor. Note that the video begins when some halos have already accumulated in the cytoplasm (00:00). Green arrowheads point to the centrosomal centrioles when discernible. Halos (red arrowheads) first accumulate in the cytoplasm (00:00–08:00) then transform into flowers (12:40; yellow arrowheads) before simultaneous centriole release (14:40). All the centrioles then migrate to the apical membrane where they grow cilia (87:00; (b)). b, Immunostaining, at the end of the time-lapse experiment shown in a, of the ependymal-specific membrane protein CD24 confirms that Cen2–GFP+ structures are docked centrioles (basal bodies) growing motile cilia. c, From Supplementary Video 4. Single z-plane images of a Cen2–GFP+ centrosomal centriole (white arrowhead) during the formation of a halo (red arrowhead). Note that the second centrosomal centriole is outside the field of observation. d, Single z-plane of centrosomal centrioles (white arrowheads) during the concomitant formation of two halos (red and green arrowheads) from one of the two centrosomal centrioles. The halo on the right (green arrowhead) detached at 04:30 then disappeared from the field. Note that the second centrosomal centriole appears in the field at 01:00 (second white arrowhead). e, From Supplementary Video 5. Three 3D-SIM z-planes (0.1 µm) showing a cytoplasmic halo with 20 procentrioles organized in a raspberry-like structure (18 Sas-6+/Cen2–GFP+ procentrioles are numbered; circles show positions of Cen2–GFP+ subunits relative to Sas-6 staining). f, Representative images of the quantifications shown in Fig. 2e (Ki67+; left). Sas-6, Cep152 and Plk4 immunoreactivity on Cen2–GFP+ mother (GT335+ primary cilium) and daughter centrosomal centrioles in cycling Ki67+ ependymal progenitors at DIV −2. ‘Fire’ LUT and fluorescence intensity profiles are shown. g, Daughter:mother centriole Sas-6 signal ratios in FoxJ1+ differentiating ependymal progenitors in P4 Cen2–GFP ependymal walls. Left: Sas-6 staining on mother (Cep164+) and daughter (Cep164−) centrosomal centrioles in halo-stage cells, their corresponding ‘fire’ LUT’s, and their fluorescence intensity profiles. Right: daughter:mother ratios of Sas-6 fluorescence intensity (error bars represent mean ± s.d.; 2.3 ± 0.9; n = 38 cells). The distribution differs significantly from a theoretical mean ratio of 1 (one sample t-test, P < 0.0001); distributions of Sas-6 ratios in differentiating progenitors in vitro (Fig. 2e) and in vivo do not differ statistically. Time in hh:mm; scale bars, 5 µm (a–b); 1 µm (c–g).
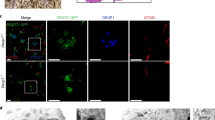
Extended Data Figure 3 Deup1 and Ccdc78 in cycling and differentiating ependymal progenitors; correlative 3D-SIM and transmission electron microscopy (3D-SIM/EM) protocol.
a, Representative images of Deup1 and Ccdc78 immunoreactivity in cycling (Ki67+) Cen2–GFP+ ependymal progenitors. b, Deup1 (left panel) and Ccdc78 (right panel) immunoreactivity on Cen2–GFP+ mother (GT335+ primary cilium) and daughter centrosomal centrioles in halo-stage cells. ‘Fire’ LUT and fluorescence intensity profiles are shown. c, Daughter:mother centriole Deup1 (5.7 ± 4.6; n = 20 cells) and Ccdc78 (3.4 ± 2; n = 20 cells) signal ratios in differentiating (FoxJ1+) ependymal progenitors at the halo stage; error bars represent mean ± s.d.. Distributions differ statistically from a theoretical mean ratio of 1 (two-tailed one sample t-test, P < 0.0001). d, Cen2–GFP/KusabiraOrange-Deup1 (K-O-Deup1) × 100 videomicroscopy image sequences showing concentric Cen2+/Deup1+ halo formation (white arrowheads) and release from one of the two centrosomal centrioles (left panel; concomitant formation of 2 halos from C2) or from the daughter centriole (right panel; Cen2–GFP occasionally enter the primary cilium, distinguishing the mother from the daughter centriole). e, Overview of the correlative 3D-SIM/EM protocol (see Methods). C1 and C2: centrosomal centrioles (when mother and daughter centrioles cannot be distinguished). Time in hh:mm; scale bars, 1 µm.
Extended Data Figure 4 Correlative 3D-SIM and transmission electron microscopy of halo formation in a Cen2-GFP ependymal progenitor (example 1).
Consecutive apical-to-basal ultra-thin EM sections (70 nm) and corresponding 3D-SIM z-projections (200 or 600 nm as indicated) are shown for both centrosomal centrioles and three cytoplasmic deuterosomes (D) in the same cell. Procentriole positions are indicated by numbers. Rectangles show the localization of the mother (green) and daughter (blue) centrioles on the preceding or following EM section. In this cell, two deuterosomes are forming from the daughter centriole. A procentriole is growing directly from the wall of the mother centriole (9). Procentrioles cut in the sagittal plane through the centre have the same appearance (2, 4, 10, 11, 13, 14, 16). In transverse sections, the procentrioles have a tube-like structure (7, 8, 9, 18). Note that the spatial resolution of 3D-SIM is not sufficient to distinguish some of the procentrioles visible by EM (1–4, 3–7, 8). Ag, aggregate of Cen2–GFP; scale bar: 0.1 µm.
Extended Data Figure 5 Correlative 3D-SIM and transmission electron microscopy of halo formation in a Cen2-GFP ependymal progenitor (example 2).
Consecutive apical-to-basal ultra-thin EM sections (70 nm) and corresponding 3D-SIM z-projections (200–700 nm as indicated) are shown for the centrosomal centrioles and 4 cytoplasmic deuterosomes (D) in the same cell. Procentriole positions are indicated by numbers. Rectangles show the localization of the mother (green) and daughter (blue) centrioles on the preceding or following EM section. In this cell, two deuterosomes are forming from the daughter centriole. A procentriole is growing directly from the wall of the daughter centriole (8). Procentrioles cut in the sagittal plane through the centre have the same appearance (2, 7, 15, 17, 19, 21). In transverse sections, the procentrioles have a tube-like structure (8, 18, 24). Note that the spatial resolution of 3D-SIM is not sufficient to distinguish some of the procentrioles visible by EM (3–5, 6, 8, 13–17, 19, 20–23). Scale bar, 0.1 µm.
Extended Data Figure 6 Formation of deuterosomes from the daughter centrosomal centriole during the halo stage and growth of procentrioles during the flower stage.
a, Formation of the first deuterosome (D) in an early differentiating ependymal progenitor (DIV 3 cell). Left: EM image showing the daughter centrosomal centriole. In contrast to the cells presented in b and c, note the low abundance of electron-dense aggregates (red arrowheads) in the cytoplasm representative of an early step of centriologenesis6. Right panels: serial EM sections spanning the centrosome of the same cell (note that the pictures have been rotated to re-orient the daughter centriole). A small deuterosome is attached to the proximal part of the centrosomal daughter centriole. Serial sections covering 2 µm of the apical portion of this cell show the absence of procentrioles and other deuterosomes. Prox., proximal. b, Serial EM sections spanning the centrosome of a differentiating ependymal progenitor showing a procentriole-loaded deuterosome attached to the proximal wall of the daughter centriole. Blue arrowheads point to procentrioles extending from the deuterosome. Yellow arrowheads point to procentrioles growing directly from the proximal part of the mother and the daughter centrioles. A rectangle and a circle indicate the position of the daughter and the mother centriole, respectively, in the preceding serial EM section. c, Left: centrosome of a cell during deuterosome formation. Note the microtubule network converging on the mother centriole. Right panels: serial EM sections spanning the centrosome. A deuterosome is attached to the proximal part of the daughter centriole. An electron-dense patch (magenta arrowhead), previously defined as centriolar precursor material in cycling cells20, can be observed on the wall of the daughter centriole adjacent to where the deuterosome contacts the centriolar wall. Blue arrowheads point to procentrioles. d, Deuterosome volume calculation protocol. Deuterosomes were outlined interactively (purple surfaces) on consecutive ultra-thin EM slices spanning the entire structure. Areas were summed for each deuterosome and multiplied by the thickness of the ultra-thin sections (0.07 µm). e, Deuterosome volume distribution relative to their localization and the number of attached procentrioles; Pearson’s correlation coefficient r = 0.67 (P = 0.001) for 20 deuterosomes from 6 cells. f, Representative TEM images of procentrioles at the halo stage (red), the flower stage (yellow), or mature centriole docked at the plasma membrane nucleating motile cilia (grey). g, Serial EM sections spanning 910 nm of the apical part of a flower-stage cell (DIV 3). Procentrioles are seen growing from both centrosomal centrioles (C1 and C2) and from deuterosomes (D1 to D11). Note the raspberry-like organization of the centrioles growing from the spherical deuterosomes. Scale bars: 0.2 µm (a); 0.1 µm (b, d), 0.5 µm (c, g).
Extended Data Figure 7 Protocols for measuring procentrioles from EM and 3D-SIM images.
a, Protocol for measuring procentrioles from EM images. Centrioles aligned parallel to the 70-nm z-slices were selected for quantification of EM width. Centriole width was quantified using a pixel intensity measurement tool placed at the distal extremities of the centrioles perpendicular to the centriolar walls. The resulting distributions of normalized pixel intensity were characterized by two minima on the y-coordinates corresponding to the centre of each of the two centriolar walls. Final plotted widths were obtained using the equation  ; where W is the absolute difference in nm between the x-coordinates of both y-axis minima. b, Protocol for measuring procentriole lengths from 3D-SIM images. Measurements were made of Cen2–GFP procentrioles, stained with Sas-6, that were parallel to the x,y plane on single 100-nm z-slices. Sas-6 staining was used to determine the longitudinal axis of the procentriole and was excluded from measurements of fluorescence intensity. Distributions of normalized Cen2–GFP fluorescence intensities were fitted using the robust nonlinear regression method, aligned, and their widths were measured to obtain a plot of procentriole lengths in nanometers. Each plotted dot corresponds to the width of the distribution curve obtained using the equation
; where W is the absolute difference in nm between the x-coordinates of both y-axis minima. b, Protocol for measuring procentriole lengths from 3D-SIM images. Measurements were made of Cen2–GFP procentrioles, stained with Sas-6, that were parallel to the x,y plane on single 100-nm z-slices. Sas-6 staining was used to determine the longitudinal axis of the procentriole and was excluded from measurements of fluorescence intensity. Distributions of normalized Cen2–GFP fluorescence intensities were fitted using the robust nonlinear regression method, aligned, and their widths were measured to obtain a plot of procentriole lengths in nanometers. Each plotted dot corresponds to the width of the distribution curve obtained using the equation  , where SD is the width of the distribution, amplitude is the height of the centre of the distribution in Y units, and mean is the x value at the centre of the distribution.
, where SD is the width of the distribution, amplitude is the height of the centre of the distribution in Y units, and mean is the x value at the centre of the distribution.
Extended Data Figure 8 Halo to flower stage transition in differentiating ependymal progenitors.
a–c, From Supplementary Video 4. Dynamics of the transformation of Cen2–GFP halos (numbered at 03:30) into flower-like structures (corresponding numbers at 08:30) after the formation of the thirteenth and last halo (red arrowhead, same cell as in Extended Data Fig. 2c). Note that procentrioles become visible on the walls of both centrosomal centrioles at the flower stage (yellow arrowheads). Centrioles detach simultaneously from both centrosome and deuterosome platforms at 10:30. C1 and C2 are centrosomal centrioles. Ag, aggregate of Cen2–GFP. d, Sas-6 immunoreactivity on 3D-SIM z-projections (left panel) or single z-slices (120 nm, right panels) of Cen2–GFP+ (green) cells at the halo and flower stages. At the halo stage, halos have accumulated in the cytoplasm and a halo is seen forming from the C1 centriole (same cell as in Fig. 2c). At the flower stage, eleven flowers are visible in the cytoplasm and procentrioles are elongating from C1 and C2 centrosomal centrioles and deuterosomes. e, Quantification of the length of Cen2–GFP+ procentrioles on 3D-SIM images of cells at the halo (red) and flower (yellow) stage. There are no significant differences among cells. The lengths of the procentrioles in flower stage-cells are statistically different from the lengths of the procentrioles in halo-stage cells (error bars represent mean ± s.d.; two-tailed unpaired t-test, ***P < 0.0001). See Extended Data Fig. 7b for quantification protocol. f, g, 3D-SIM z-slices (120 nm) showing the immunolocalization of Sas-6, Deup1, Cep152, Plk4, Stil, Cpap and CP110 in Cen2GFP+ halo (f) and flower (g) procentrioles. h, Immunostaining of P4 ependymal walls showing that Sas-6 immunoreactivity disappears from Cep-164+ centrioles when they begin to grow motile cilia (GT335+). Time in hh:mm; scale bars, 1 µm.
Extended Data Figure 9 Ciliary mutant analyses in vitro and centriole amplification in mouse trachea in vivo.
a–d, Ciliary mutant analyses in vitro. a, GT335 and Sas-6 immunoreactivity in a Cen2–GFP+ cycling ependymal progenitor (DIV −2; Ki67+) and a differentiating progenitor at the halo stage (DIV3). Arrowheads point to halos. b, Quantification of primary cilium length in cycling (1 ± 0.4 µm; n = 25 cells) and differentiating progenitors (3.2 ± 0.6 µm; n = 32 cells) showing a significant elongation at the halo stage (error bars represent mean ± s.d.; two-tailed Mann–Whitney U-test, ***P < 0.0001). c, d, The absence of a primary cilium in differentiating ependymal progenitor cells from Kif3a cKO mutants does not affect Sas-6 asymmetry. c, Representative Sas-6 staining on Cen2–GFP+ mother (Cep164+) and daughter centrosomal centrioles in control (left panel) and Kif3a cKO (right panel) halo-stage cells. Second and third columns show, respectively, the corresponding ‘fire’ LUT signal intensities and fluorescence intensity profiles. d, Daughter:mother centriole Sas-6 signal ratios in differentiating (FoxJ1+) control (2.4 ± 1.5; n = 30 cells) and Kif3a cKO (2.3 ± 1.3; n = 29 cells) ependymal progenitor cells at the halo stage. The distributions are not statistically different, but both differ significantly from a theoretical mean ratio of 1 (error bars represent mean ± s.d.; two-tailed one sample t-test, P < 0.0001; ns, not significant). e–k, Centriole amplification in mouse trachea in vivo. e, Bright-field image showing a typical cranio-dorsal region of the E17-P0 trachea whole mounts analysed in (f–k). f, Merged images of Sas-6-immunostainined Cen2–GFP+ tracheal multiciliated progenitors showing that the steps of centriole amplification are the same as in brain ependymal walls; the stainings in the white and yellow boxes are shown respectively in the upper and lower panels of g. g, Single 0.24 µm z-slices of the boxed regions in f: the centrosome images show Sas-6 negative centrosomal centrioles; the halo images show that halos arise from the wall of a centrosomal centriole (upper panels) and accumulate in the cytoplasm (lower panels); the flower images show procentrioles on a centrosomal centriole (centriolar pathway; upper panels) and hollow Sas-6+ flowers (deuterosome pathway; lower panels); the basal body images show mature docked Sas-6-negative basal bodies. White arrowheads point to centrosomal centrioles in f and g. h, Sas-6 staining on Cen2–GFP+ mother (ninein+) and daughter centrosomal centrioles in halo-stage cells in trachea epithelium. Second and third columns show, respectively, the corresponding ‘fire’ LUT signal intensities and fluorescence intensity profiles of the same markers. i, Daughter:mother centriole Sas-6 signal ratios (2.3 ± 1.3; n = 36 cells) in halo-stage cells in E17–P0 trachea. The distribution differs statistically from a theoretical mean ratio of 1 (error bars represent mean ± s.d.; two-tailed one sample t-test, P < 0.0001); Sas-6 halo-stage daughter/mother centriole ratios are similar in ependymal walls (Extended Data Fig. 2g) and trachea epithelium. j, Upper panels: 0.96 µm z-stacked images of a typical Sas-6/ninein immunoreactivity on E17–P0 Cen2–GFP trachea showing that halo structures form exclusively on the daughter centriole (n = 12 cells). Lower panels: 0.96 µm z-stacked images of a typical Deup1/ninein immunoreactivity on E17–P0 Cen2–GFP trachea showing accumulation of Deup1 on the daughter centriole. k, Immunostaining of P0 trachea showing that Sas-6 immunoreactivity disappears from basal bodies when they begin to grow GT335+ motile cilia. Scale bars, 1 µm.
Extended Data Figure 10 Hypothetical model of centriole amplification through deuterosome formation.
a, An unknown mechanism leads to Deup1 and Ccdc78 accumulation at the daughter centriole. b, Binding of Cep152 and recruitment of Plk4 (refs 7,8) activate nucleation of the first procentriole. c, Accumulation continues from the same active site and displace the first procentriole. d, The second procentriole arises from the new complex. e–g, The process continues until the deuterosome detaches from the centriolar wall. g, The process begins again to form another deuterosome and nucleate new procentrioles. During deuterosome formation (halo stage), procentrioles remain latent at an early stage of their biogenesis. Several deuterosomes can form concomitantly from different active sites on the daughter centriole proximal wall. h, The cycle continues until centriole growth is triggered and procentrioles grow from the walls of both centrosomal centrioles and deuterosomes (flower stage). See Fig. 4i, j and Extended Data Fig. 8f, g for 3D-SIM localization of structural and regulatory proteins depicted here.
Supplementary information
Cen2-GFP live imaging during centriole amplification in ependymal cell (example 1)
Apical (upper panel) and side (lower panel) views of a time-lapse sequence showing Cen2-GFP dynamics (63X magnification, ∆t= 40 minutes) during the maturation of an ependymal progenitor in vitro. Note that halos appear within the centrosome region (14:00-30:40) and transform into flower-like structures (31:20-34:00) before the simultaneous release and apical docking of all the centrioles (34:00-72:40). White arrowheads point to the centrosomal centrioles of the ependymal progenitor when discernible. See Fig. 1(d-h). Time is in hh:mm. Scale bar: 2μm. (MOV 4689 kb)
Cen2-GFP live imaging during centriole amplification in ependymal cell (example 2)
Apical (upper panel) and side (lower panel) views of a time-lapse sequence showing Cen2-GFP dynamics (63X magnification, ∆t= 40 minutes) during the maturation of an ependymal progenitor in vitro, starting from the halo formation stage (00:00-04:00). Halos transform into flower-like structures (04:40-12:40) prior to the simultaneous release and apical docking of all the centrioles (12:40-32:40). Cd24 staining at 87:20 shows that basal bodies have nucleated cilia. White arrowheads indicate the centrosomal centrioles of the ependymal progenitor when discernible. See Extended Data Fig. 2(a-b). Time in hh:mm. Scale bar: 2μm. (MOV 2570 kb)
Cen2-GFP live imaging during halo formation (example 1)
Apical view of a time-lapse sequence showing Cen2-GFP dynamics (100X magnification, ∆t= 30 minutes) during the process of halo formation. Note that 6 halos have already formed and are numbered in the cytoplasm. White arrowheads indicate centrosomal centrioles. Red arrowhead indicates the 7th and last halo formed from a centrosomal centriole (01:00-04:30). After the last halo is released, the 7 halos transform into flowers (numbered at 12:30). Centrioles become visible on the walls of both centrosomal centrioles (yellow arrowheads). See Fig. 2a for single 0.7 μm z-planes of halo formation. Time in hh:mm. Scale bar: 2μm. (MOV 13563 kb)
Cen2-GFP live imaging during halo formation (example 2)
Twelve numbered halos have already formed. White arrowheads indicate centrosomal centrioles. Red arrowhead indicates the 13th and last halo formed from a centrosomal centriole (01:00-08:30). All halos then transform simultaneously into flowers (numbered at 08:30). Centrioles become visible on the wall of both centrosomal centrioles (yellow arrowheads). Centrioles detach simultaneously from both centrosome and deuterosome platforms from 10:30. Note that at 04:30, the halo is going under the second centrosomal centriole. Centrin aggregates are occasionally observed in Cen2-GFP cells. See Extended Data Fig. 2c for single z-planes of halo formation. Time in hh:mm. Scale bar: 2μm. (MOV 1835 kb)
3D-SIM z-planes of a halo
3D-SIM z-planes (0.1μm) spanning a halo and showing the raspberry-like organization of 20 procentrioles (green: Cen2-GFP, red: SAS-6). Dashed circles (left) outline the Cen2+ subunits and dashed rectangles (right) outline the Cen2+/Sas-6+ procentrioles that are oriented parallel to z-plane. z labeling is in μm. (MOV 121 kb)
Cen2-GFP/Kusabira-Orange-Deup1 live imaging during halo formation
Time-lapse sequence of the Cen2-GFP/KusabiraOrange-Deup1 dynamics (100X magnification, Δt= 30 minutes) during centriole amplification showing that halos and flowers display a Deup1+ core. Ten halos are already cytoplasmic, some of which are superimposed due to z-stacking. White arrowheads indicate centrosomal centrioles. Magenta arrowhead indicates the 11th and last halo formed from a centrosomal centriole (00:00-01:30). After the last halo is formed, all the halos transform simultaneously into flower-like structures by 05:00. Cyan arrowheads indicate procentrioles that become visible on the walls of both centrosomal centrioles. Centrin aggregates are occasionally observed in Cen2-GFP expressing cells. Time in hh:mm. Scale bar: 2μm. (MOV 304 kb)
Cen2-GFP live imaging during centriole detachment from centrosomal centrioles and deuterosomes
Cen2-GFP time-lapse imaging (100X magnification, Δt= 30 minutes) at the end of the flower stage showing the synchronized release of procentrioles (t=00:00-03:00) from both centrosomal centrioles and deuterosomes (¯t=2.5±0.85h in 9 cells from 5 independent experiments). Released centrioles have migrated to the apical membrane by 20:30. White arrowheads point to the centrosomal centrioles when discernible. Halos appearing at -01:30 in the lower right corner and flowers appearing at 11:00 are from neighboring cells. Time in hh:mm. Scale bar: 2μm. (MOV 2219 kb)
Rights and permissions
About this article
Cite this article
Al Jord, A., Lemaître, AI., Delgehyr, N. et al. Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature 516, 104–107 (2014). https://doi.org/10.1038/nature13770
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13770
This article is cited by
-
RBL2 represses the transcriptional activity of Multicilin to inhibit multiciliogenesis
Cell Death & Disease (2024)
-
Appearing and disappearing acts of cilia
Journal of Biosciences (2023)
-
Ependymoma associated protein Zfta is expressed in immature ependymal cells but is not essential for ependymal development in mice
Scientific Reports (2022)
-
Astroblastomas exhibit radial glia stem cell lineages and differential expression of imprinted and X-inactivation escape genes
Nature Communications (2022)
-
Dynamics of centriole amplification in centrosome-depleted brain multiciliated progenitors
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.