Abstract
The reprogramming of epigenetic states in gametes and embryos is essential for correct development in plants and mammals1. In plants, the germ line arises from somatic tissues of the flower, necessitating the erasure of chromatin modifications that have accumulated at specific loci during development or in response to external stimuli. If this process occurs inefficiently, it can lead to epigenetic states being inherited from one generation to the next2,3,4. However, in most cases, accumulated epigenetic modifications are efficiently erased before the next generation. An important example of epigenetic reprogramming in plants is the resetting of the expression of the floral repressor locus FLC in Arabidopsis thaliana. FLC is epigenetically silenced by prolonged cold in a process called vernalization. However, the locus is reactivated before the completion of seed development, ensuring the requirement for vernalization in every generation. In contrast to our detailed understanding of the polycomb-mediated epigenetic silencing induced by vernalization, little is known about the mechanism involved in the reactivation of FLC. Here we show that a hypomorphic mutation in the jumonji-domain-containing protein ELF6 impaired the reactivation of FLC in reproductive tissues, leading to the inheritance of a partially vernalized state. ELF6 has H3K27me3 demethylase activity, and the mutation reduced this enzymatic activity in planta. Consistent with this, in the next generation of mutant plants, H3K27me3 levels at the FLC locus stayed higher, and FLC expression remained lower, than in the wild type. Our data reveal an ancient role for H3K27 demethylation in the reprogramming of epigenetic states in plant and mammalian embryos5,6,7.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Feng, S., Jacobsen, S. E. & Reik, W. Epigenetic reprogramming in plant and animal development. Science 330, 622–627 (2010)
Paszkowski, J. & Grossniklaus, U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 14, 195–203 (2011)
Becker, C. et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249 (2011)
Schmitz, R. J. et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334, 369–373 (2011)
Mansour, A. A. et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 488, 409–413 (2012)
Canovas, S., Cibelli, J. B. & Ross, P. J. Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development. Proc. Natl Acad. Sci. USA 109, 2400–2405 (2012)
Zhao, W. et al. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell 152, 1037–1050 (2013)
Johanson, U. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347 (2000)
Sheldon, C. C. et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 (1999)
Gendall, A. R., Levy, Y. Y., Wilson, A. & Dean, C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535 (2001)
Song, J., Angel, A., Howard, M. & Dean, C. Vernalization: a cold-induced epigenetic switch. J. Cell Sci. 125, 3723–3731 (2012)
De Lucia, F., Crevillen, P., Jones, A. M. E., Greb, T. & Dean, C. A PHD–polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl Acad. Sci. USA 105, 16831–16836 (2008)
Sheldon, C. C. et al. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl Acad. Sci. USA 105, 2214–2219 (2008)
Choi, J. et al. Resetting and regulation of FLOWERING LOCUS C expression during Arabidopsis reproductive development. Plant J. 57, 918–931 (2009)
Mylne, J., Greb, T., Lister, C. & Dean, C. Epigenetic regulation in the control of flowering. Cold Spring Harb. Symp. Quant. Biol. 69, 457–464 (2004)
Lu, F., Cui, X., Zhang, S., Jenuwein, T. & Cao, X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nature Genet. 43, 715–719 (2011)
Noh, B. et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16, 2601–2613 (2004)
Roeder, A. H. K. & Yanofsky, M. F. Fruit development in Arabidopsis. Arabidopsis Book 4, e0075 (2006)
Bastow, R. et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167 (2004)
Ietswaart, R., Wu, Z. & Dean, C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28, 445–453 (2012)
Slotkin, R. K. et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472 (2009)
Mosher, R. A. et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460, 283–286 (2009)
Swiezewski, S. et al. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl Acad. Sci. USA 104, 3633–3638 (2007)
Alexandre, C. M. & Hennig, L. FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59, 1127–1135 (2008)
Kim, D.-H. & Sung, S. Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25, 454–469 (2013)
Ingouff, M. et al. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 20, 2137–2143 (2010)
Surani, M. A., Hayashi, K. & Hajkova, P. Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 (2007)
Crevillén, P., Sonmez, C., Wu, Z. & Dean, C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 32, 140–148 (2013)
Gu, X., Jiang, D., Wang, Y., Bachmair, A. & He, Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57, 522–533 (2009)
Jiang, D., Gu, X. & He, Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21, 1733–1746 (2009)
Oñate-Sánchez, L. & Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1, 93 (2008)
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008)
Stein, L. D. et al. The generic genome browser: a building block for a model organism system database. Genome Res. 12, 1599–1610 (2002)
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988)
Earley, K. W. et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 (2006)
Angel, A., Song, J., Dean, C. & Howard, M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108 (2011)
Acknowledgements
We thank Dean laboratory members and A. Surani for discussions. The Dean laboratory is supported by the UK Biotechnology and Biological Sciences Research Council grants BB/G009562/1 and BB/C517633/1 and by a European Research Council Advanced Investigator grant (233039 ENVGENE). The Cao laboratory is supported by National Basic Research Program of China grants 2013CB967300 and 2011CB915400 and by the National Natural Science Foundation of China grant 31271363.
Author information
Authors and Affiliations
Contributions
P.C. and C.D. designed the research. P.C., H.Y., X. Cui, C.G. and Q.Q. performed experiments. M.T. conducted deep sequencing data analysis. X. Cao contributed new reagents and analytical tools. P.C., H.Y. and C.D. analysed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
C.D. holds stock in Mendel Biotechnology.
Extended data figures and tables
Extended Data Figure 1 Screening for mutants with impaired epigenetic reprogramming of FLC.
a, We started with a population of ethylmethane sulphonate (EMS)-mutagenized A. thaliana Ler plants carrying an FLC::LUC translational fusion and an active FRI transgene. We screened for mutants that were early flowering (as a result of low FLC expression) in the generation following vernalization but that did not flower early (and whose FLC expression was almost normal) without vernalization. b, To discriminate between early flowering and resetting mutants, early flowering M2 plants were backcrossed to the parental line, and the F2 phenotype was evaluated without vernalization. Those plants showing no early flowering segregants were considered to be resetting mutants. Superscript characters denote whether the plant was vernalized in the previous generation.
Extended Data Figure 2 Characterization of the first resetting mutant.
a, The first resetting mutation was found to be recessive. F1 plants were generated from a cross between the mutant in the generation following vernalization and the parental wild-type line. Flowering time was assayed as total leaf number under non-vernalized long-day conditions. The data are presented as the mean + s.e.m., n = 8. b, The earlier flowering time of the mutant in the generation following vernalization was stable for at least three generations without vernalization. The data are presented as the mean + s.e.m., n = 10.
Extended Data Figure 3 The elf6-5 SNP is linked to the resetting of FLC expression.
Histogram showing the relationship between FLC::LUC levels in the reproductive organs of vernalized plants and the elf6-5 SNP (n = 154).
Extended Data Figure 4 FLC expression levels in the null elf6-3 T-DNA insertion allele.
a, The elf6-3 mutant expresses less FLC than the Col wild type. b, The null elf6-3 allele suppresses the high FLC expression induced by FRI before vernalization. This pre-vernalization phenotype of plants carrying the null allele precludes observation of the role of ELF6 during FLC resetting after vernalization. All graphs show 10-day-old non-vernalized seedlings. The data are presented as the mean + s.e.m., n = 3.
Extended Data Figure 5 siRNA production in elf6-5 mutants.
The production of specific siRNAs associated with the epigenetic reactivation of transposable elements is not affected in elf6-5 mutants. Total RNA was extracted from vernalized mature siliques, and the detection of siRNAs was performed as described in Methods.
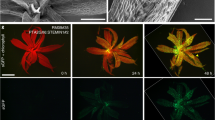
Extended Data Figure 6 ELF6 has no H3K4me, H3K9me or H3K36me demethylase activity in an N. benthamiana transient assay.
Overexpression of a yellow fluorescent protein (YFP)–ELF6 fusion protein, using the wild-type ELF6 sequence had no effect on H3K4me3, H3K9me2 or H3K36me3 methylation. Histone methylation was visualized by immunostaining with polyclonal rabbit modification-specific antibodies followed by Alexa-Fluor-555-conjugated goat anti-rabbit antibody (red; right). The nuclei of transfected cells were visualized by the YFP signal (green; centre). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (blue; left). Arrows indicate the nuclei of transfected cells.
Extended Data Figure 7 H3K27me3 accumulation at the FLC locus.
a, Schematic representation of the FLC locus and the regions analysed in the ChIP assays. b, H3K27me3 levels at FLC in Ler (FRI) seedlings grown without vernalization (seedling NV), 7 days after vernalization (seedling VER) and in siliques from vernalized seedlings (siliques VER). The data are presented as the mean + s.d., n = 2.
Rights and permissions
About this article
Cite this article
Crevillén, P., Yang, H., Cui, X. et al. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590 (2014). https://doi.org/10.1038/nature13722
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13722
This article is cited by
-
RWP-RK Domain 3 (OsRKD3) induces somatic embryogenesis in black rice
BMC Plant Biology (2023)
-
Distinct chromatin signatures in the Arabidopsis male gametophyte
Nature Genetics (2023)
-
A robust mechanism for resetting juvenility during each generation in Arabidopsis
Nature Plants (2022)
-
Epigenetic repression and resetting of a floral repressor, FLC, in the life cycle of winter-annual Arabidopsis
Plant Biotechnology Reports (2022)
-
Plant synthetic epigenomic engineering for crop improvement
Science China Life Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.