Abstract
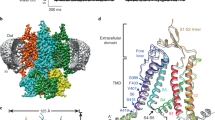
Cys-loop receptors are neurotransmitter-gated ion channels that are essential mediators of fast chemical neurotransmission and are associated with a large number of neurological diseases and disorders, as well as parasitic infections1,2,3,4. Members of this ion channel superfamily mediate excitatory or inhibitory neurotransmission depending on their ligand and ion selectivity. Structural information for Cys-loop receptors comes from several sources including electron microscopic studies of the nicotinic acetylcholine receptor5, high-resolution X-ray structures of extracellular domains6 and X-ray structures of bacterial orthologues7,8,9,10. In 2011 our group published structures of the Caenorhabditis elegans glutamate-gated chloride channel (GluCl) in complex with the allosteric partial agonist ivermectin, which provided insights into the structure of a possibly open state of a eukaryotic Cys-loop receptor, the basis for anion selectivity and channel block, and the mechanism by which ivermectin and related molecules stabilize the open state and potentiate neurotransmitter binding11. However, there remain unanswered questions about the mechanism of channel opening and closing, the location and nature of the shut ion channel gate, the transitions between the closed/resting, open/activated and closed/desensitized states, and the mechanism by which conformational changes are coupled between the extracellular, orthosteric agonist binding domain and the transmembrane, ion channel domain. Here we present two conformationally distinct structures of C. elegans GluCl in the absence of ivermectin. Structural comparisons reveal a quaternary activation mechanism arising from rigid-body movements between the extracellular and transmembrane domains and a mechanism for modulation of the receptor by phospholipids.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Thompson, A. J., Lester, H. A. & Lummis, S. C. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 43, 449–499 (2010)
Sine, S. M. End-plate acetylcholine receptor: structure, mechanism, pharmacology, and disease. Physiol. Rev. 92, 1189–1234 (2012)
Corringer, P. J. et al. Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure 20, 941–956 (2012)
Boatin, B. A. & Richards, F. O. J. Control of onchocerciasis. Adv. Parasitol. 61, 349–394 (2006)
Unwin, N. Refined structure of the nicotinic acetylcholine receptor. J. Mol. Biol. 346, 967–989 (2005)
Brejc, K. et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 (2001)
Hilf, R. & Dutzler, R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 (2009)
Hilf, R. & Dutzler, R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008)
Bocquet, N. et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 (2009)
Sauguet, L. et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl Acad. Sci. USA 111, 966–971 (2014)
Hibbs, R. E. & Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011)
Miyazawa, A., Fujiyoshi, Y. & Unwin, N. Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955 (2003)
Bali, M. & Akabas, M. H. The location of a closed channel gate in the GABBA receptor channel. J. Gen. Physiol. 129, 145–159 (2007)
Revah, F. et al. Mutations in the channel domain alter desensization of a neuronal nicotinic receptor. Nature 353, 846–849 (1991)
Labarca, C. et al. Channel gating governed symmetrically by conserved leucine residues in the M2 domain of nicotinic receptors. Nature 376, 514–516 (1995)
Filatov, G. N. & White, M. M. The role of conserved leucines in the M2 domain of the acetylcholine receptor in channel gating. Mol. Pharmacol. 48, 379–384 (1995)
Yoluk, O., Brömstrup, T., Bertaccini, E. J., Trudell, J. R. & Lindahl, E. Stabilization of the GluCl ligand-gated ion channel in the presence and absence of ivermectin. Biophys. J. 105, 640–647 (2013)
Barrantes, F. J. Structural basis for lipid modulation of nicotinic acetylcholine receptor function. Brain Res. Brain Res. Rev. 47, 71–95 (2004)
daCosta, C. J., Dey, L., Therien, J. P. & Baenziger, J. E. A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nature Chem. Biol. 9, 701–707 (2013)
Lee, W. Y., Free, C. R. & Sine, S. M. Nicotinic receptor interloop proline anchors beta1-beta2 and Cys loops in coupling agonist binding to channel gating. J. Gen. Physiol. 132, 265–278 (2008)
Celie, P. H. et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914 (2004)
Hansen, S. B. et al. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 24, 3635–3646 (2005)
Damle, V. N. & Karlin, A. Effects of agonists and antagonists on the reactivity of the binding site disulfide in acetylcholine receptor from Torpedo californica. Biochemistry 19, 3924–3932 (1980)
Lynagh, T. & Lynch, J. W. A glycine residue essential for high ivermectin sensitivity in Cys-loop ion channel receptors. Int. J. Parasitol. 40, 1477–1481 (2010)
Velisetty, P., Chalamalasetti, S. V. & Chakrapani, S. Structural basis for allosteric coupling at the membrane-protein interface in Gloeobacter violaceus ligand-gated ion channel (GLIC). J. Biol. Chem. 289, 3013–3025 (2014)
Reeves, D. C., Jansen, M., Bali, M., Lemster, T. & Akabas, M. H. A role for the β1-β2 loop in the gating of 5HT3 receptors. J. Neurosci. 25, 9358–9366 (2005)
Calimet, N. et al. A gating mechanism of pentameric ligand-gated ion channels. Proc. Natl Acad. Sci. USA 110, E3987–E3996 (2013)
Purohit, P., Gupta, S., Jadey, S. & Auerbach, A. Functional anatomy of an allosteric protein. Nature Commun. 4, 2984 (2013)
Gleitsman, K. R., Lester, H. A. & Dougherty, D. A. Probing the role of backbone hydrogen bonding in a critical beta sheet of the extracellular domain of a cys-loop receptor. ChemBioChem 10, 1385–1391 (2009)
Lee, W. Y. & Sine, S. M. Principle pathway coupling agonist binding to channel gating in nicotinic receptors. Nature 438, 243–247 (2005)
Gourdon, P. et al. HiLiDe–Systematic approach to membrane protein crystallization in lipid and detergent. Cryst. Growth Des. 11, 2098–2106 (2011)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Hanson, M. A. et al. Crystal structure of a lipid-G protein-coupled receptor. Science 335, 851–855 (2012)
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007)
Cowtan, K. D. & Zhang, K. Y. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 72, 245–270 (1999)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Painter, J. & Merritt, E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 (2006)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Sauguet, L. et al. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 32, 728–741 (2013)
DeLano, W. L. The PyMOL molecular graphics system. http://www.pymol.org (DeLano Scientific, 2002)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Samsom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996)
Poornam, G. P., Matsumoto, A., Ishida, H. & Hayward, S. A method for the analysis of domain movements in large biomolecular complexes. Proteins 76, 201–212 (2009)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Hart, H. E. & Greenwald, E. B. Scintillation proximity assay (SPA) - a new method of immunoassay. Direct and inhibition mode detection with human albumin and rabbit antihuman albumin. Mol. Immunol. 16, 265–267 (1979)
Acknowledgements
We thank all staff of beamline 24-ID-C at the Advanced Photon Source. We thank L. Vaskalis and H. Owen for help in figure and manuscript preparation, respectively. D. Cawley at the Vaccine and Gene Therapy Institute, OHSU, provided the monoclonal antibody. We appreciate discussions with Gouaux laboratory members. This work was supported by a postdoctoral fellowship (Forschungsstipendium AL 1725-1/1) from the Deutsche Forschungsgemeinschaft to T.A. and an individual NIH National Research Service Award (F32NS061404) to R.E.H; E.G. is supported by the NIH and is an investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
T.A., R.E.H. and S.B. performed the experiments and T.A., R.E.H. and E.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Packing of two pentameric GluCl–Fab complexes in the asymmetric unit of the C2 unit cell.
a, Asymmetric unit of apo GluClcryst–Fab complex seen in the plane of the membrane. The receptor is coloured by subunit and the Fab is displayed in grey. b, Complex from a rotated by 90°. c, d, The two GluClcryst–Fab complexes with POPC (shown as yellow spheres) seen parallel (c) and perpendicular (d) to the putative membrane plane.
Extended Data Figure 2 Electron density maps for the apo GluCl structure.
Shown are electron density maps calculated using 2Fo − Fc coefficients and contoured at 1.5σ with the α-carbon trace for approximately opposing subunits P and R seen in the plane of the membrane (a), the pore with the gating Leu 254 seen from the extracellular side (b), and the interface between the β1–β2 loop from the extracellular domain and the M2–M3 loop from the transmembrane domain with Val 45 and Pro 268 shown as sticks (c).
Extended Data Figure 3 Comparison of apo structures for GluCl, ELIC and GLIC.
a, d, Two approximately opposing subunits (P and R or A and D) are shown for the apo state of GluCl (blue) and ELIC (PDB 2VL0; yellow) (a), and for the apo state of GluCl (blue) and apo state of GLIC (PDB 4NPQ; green) (d) after superimposing all α-carbon atoms of one subunit. Indicated are distances between α-carbon atoms of Ser 265, Leu 254 and Pro 243 for GluCl, Asn 250, Leu 239 and Pro 228 for ELIC and Thr 244, Ile 233 and Glu 222 for GLIC (from top). b, e, Interactions of the β1–β2 loop and the M2–M3 loop in apo GluCl (blue) and ELIC (yellow) (b) and GLIC (green) (e), respectively. Key residues are shown in stick representation with the distances between them indicated. For comparison the ivermectin-bound structure is shown in grey. Panels c and f show the ion channel pore with the proposed gate as seen from the extracellular side with the same colour coding for the proteins as before.
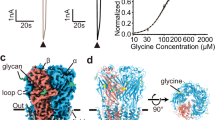
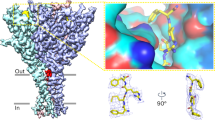
Extended Data Figure 4 Lipid binding to GluCl.
a, View of one GluCl pentamer from the extracellular side together with contours from an ‘omit’ style electron density map computed using Fo − Fc coefficients and phases from coordinates where atoms associated with POPC molecules were omitted from the structure factor computation. The electron density map is shown as blue mesh and is contoured at 1.5σ. Prominent electron density features are visible between transmembrane domains of adjacent subunits although the strength and continuity of the density varies from subunit to subunit. b, Close-up view of one putative electron density feature with POPC drawn as stick representation and viewed in the plane of the membrane. c, Radioligand competition binding experiment using GluClcryst, [3H]-ivermectin and cold POPS yields a Ki for POPS of 167 μM (95% confidence interval: 129–216 μM). Shown is a representative binding curve. d, Saturation binding experiment with [3H]-l-glutamate GluClcryst-Nano15 yields a Kd of ∼1.12 μM (95% confidence interval: 0.75–1.67 μM) in the presence of 3 mM POPS. e, Radioligand saturation binding using [3H]-ivermectin and the GluClcryst construct. Fitting the data to the Hill equation yields an EC50 of 18.5 nM (95% confidence interval of 11.1–30.7 nM) and an n = 0.76 ± 0.16. For panels c–e, experiments were carried out three separate times, with experiments done in triplicate.
Extended Data Figure 5 Superpositions of a single subunit.
a, Stereo image of the α-carbon trace of the superpositions of the extracellular domain (residues 1–211) of a single subunit for apo (blue), ivermectin- (grey) and POPC-bound (orange) conformations. View parallel to the plane of the membrane with key features indicated. b, Same view of a GluCl subunit in the three different conformations coloured as in a after superposition of the transmembrane domain (residues 212–342) to display differences in the extracellular domain. c, r.m.s.d. for the superpositions shown in panels a and b and whole subunits.
Extended Data Figure 6 Conformational changes within a subunit.
a–c, Views of the transmembrane domain seen from the extracellular side after superposition of the extracellular domain (residues 1–211) for the apo (blue) and the ivermectin-bound (grey) complex (a), for the apo and POPC-bound (orange) complexes (b) and for the POPC- and ivermectin-bound structures (c). Panels d–f illustrate relative conformational changes within a subunit when the transmembrane domains are superimposed, using residues 212–342 and the same colour coding as in panels a–c. g–i, Views from the extracellular side onto the extracellular domain after superimposing the transmembrane domain of the apo and ivermectin-bound conformation (g), the apo and POPC-bound structures (h) and the POPC- and ivermectin-bound complexes (i). The tilt and twist axes between transmembrane domain and extracellular domain are indicated by arrows as marked in the figure panels.
Extended Data Figure 7 Ivermectin and POPC alter subunit–subunit contacts in the transmembrane and extracellular domains.
Views of two neighbouring subunits of GluCl seen parallel to the plane of the membrane after superposition of the transmembrane domain of the (−)-subunit. a, Superposition of apo (blue) and ivermectin-bound (grey/white) structures. Ivermectin is shown in pale green as stick representation. b, Superposition of the apo (blue) and POPC-bound (brown/orange) complexes with POPC shown as yellow sticks. c, Superposition of POPC- (brown/orange) and ivermectin-bound (grey/white) conformations with POPC and ivermectin as yellow and pale green sticks, respectively.
Extended Data Figure 8 Coupling between extracellular and transmembrane domain.
a, Superposition of the transmembrane domain of the (−)-subunits for the apo (blue) and POPC-bound (brown) states shows the relative movement of the transmembrane domains and the M2–M3 loop of the (+)-subunit. b, Same superposition as panel a, viewed approximately parallel to the membrane, showing the coupled movement of the β1–β2 loop and the M2–M3 loop, with displacement of key residues Val 45, Pro 268 and Ile 273 indicated. Panels c and d show the same views for superposition of the POPC- (brown) and ivermectin-bound (grey) states. e–h, Illustration of key residues forming hydrogen bonds and a salt bridge that connect the transmembrane domains M3 and M1 with the Cys-loop in the extracellular domain in the ivermectin-bound state (grey) (e, f) and POPC-bound state (orange) (g, h). Shown are two views approximately in the plane of the membrane.
Extended Data Figure 9 Conformational changes in the GluCl pentamer.
a–c, Views of two opposing subunits as seen in the plane of the membrane for apo-closed (a), ivermectin-open (b) and apo-POPC (c) states. Leu 254 and Pro 243 are highlighted as sticks. Indicated are distances between α-carbon atoms of Thr 11, Ser 265 and Pro 243 (from top). d–f, Views of the transmembrane domains of the pentamer seen from the extracellular side along the pore axis for apo-closed (d), ivermectin- (e) and POPC-bound (f) conformations. For comparison the apo-closed state is shown for one subunit with the ivermectin- (e) and POPC-bound (f) states. g–i, Top views of the extracellular domains of the GluCl pentamer of the apo (g), ivermectin- (h) and POPC-bound (i) states. In panels h, i, the apo pentamer was superimposed on the ivermectin- and POPC-bound states using all atoms of the pentamer, and one subunit of the apo state (blue) is shown for comparison. We note that despite the α1-helices moving towards the pore axis, the pore diameter in the extracellular domain increases in the ivermectin- and POPC-bound conformations as the lower parts of the extracellular domain move away from the pore centre.
Supplementary information
Morph from apo to ivermectin-bound conformation of the pentamer
This video illustrates the transition of GluCl from the apo to the ivermectin-bound state from different perspectives. Key residues as pointed out in the text are shown as sticks. (MP4 12924 kb)
Morph from apo to POPC-bound conformation of the pentamer
Shown is the transition of the GluCl pentamer from the apo to the POPC-bound structure from different perspectives. Key residues are shown as sticks. (MP4 12340 kb)
Rights and permissions
About this article
Cite this article
Althoff, T., Hibbs, R., Banerjee, S. et al. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 (2014). https://doi.org/10.1038/nature13669
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13669
This article is cited by
-
The modes of action of ion-channel-targeting neurotoxic insecticides: lessons from structural biology
Nature Structural & Molecular Biology (2023)
-
Molecular encoding and synaptic decoding of context during salt chemotaxis in C. elegans
Nature Communications (2022)
-
A Pharmacological Perspective on Plant-derived Bioactive Molecules for Epilepsy
Neurochemical Research (2021)
-
Nematicidal and insecticidal activities of halogenated indoles
Scientific Reports (2019)
-
A lipid site shapes the agonist response of a pentameric ligand-gated ion channel
Nature Chemical Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.