Abstract
Replying to S. Shohat & S. Shifman Nature 512, 10.1038/nature13583 (2014)
Shohat and Shifman’s analysis1 indicates that long autism spectrum disorder (ASD) genes are overrepresented in the SFARI Gene/AutDB database (as of 11 December 2013) owing to the discovery method. We agree with their analysis and with the need to consider the strength of evidence behind each candidate gene. When our study was underway2, SFARI Gene provided the only comprehensive list of autism candidate genes with confidence values. Subsequent to our publication, more genes have been added to this database and scored, highlighting the rapid pace of advances in the ASD field and the changing confidence behind each ASD gene.
Similar content being viewed by others
Main
We agree with Shohat and Shifman that the proportion of ASD genes that are long may drop as more ASD genes are identified. We did not account for how the discovery method used to identify a given ASD candidate, be it based on copy number variant (CNV) or single nucleotide variant (SNV), might affect average gene length in our study. However, it is also undeniable that many long genes are considered candidates in ASD pathology, such as NRXN1 and CNTNAP2. Moreover, our mechanistic findings are not in dispute. Indeed, three other groups came to the same conclusion as us—that topoisomerases preferentially facilitate expression of long genes3,4,5. Our study demonstrates an essential role for topoisomerases in transcriptional elongation of long neuronal genes and suggests a critical role for these enzymes in neurodevelopmental disorders like autism.
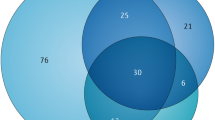
Shohat and Shifman1 also suggest that the SFARI Gene database contains many genes with weak links to ASD pathology. In our study, we did not rank genes as stronger or weaker ASD candidates, and treated all equally. However, when the degree of evidence behind each candidate is taken into account, using the gene scoring module in SFARI Gene 2.06 (as of 1 April 2014), it remains clear that numerous strong ASD candidate genes are very long (>200 kilobases). Thus, we feel our conclusion linking topoisomerases and gene length with autism is still warranted, but this remains to be tested more rigorously pending in vivo studies with animal models and additional human genetic studies.
Future studies are likely to validate additional long genes as strong ASD candidates. For example, NRXN3 and CNTN5 (1.5 and 1.3 megabase, respectively) are not yet scored in SFARI Gene, yet these genes are deleted in patients with ASD7,8 and both are significantly reduced in topotecan-treated neurons2.
Ultimately, we agree that making conclusions about the nature of ASD genes is complicated by factors like the ones Shohat and Shifman describe1 as well as by the evolving knowledge of autism genetics. Regardless, we identified a transcriptional mechanism that affects the expression of long genes, a number of which are currently classified as strong ASD candidates.
References
Shohat, S. & Shifman, S. Bias towards large genes in autism. Nature 512, http://dx.doi.org/10.1038/nature13583 (2014)
King, I. F. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 (2013)
Solier, S. et al. Transcription poisoning by topoisomerase I is controlled by gene length, splice sites, and miR-142–3p. Cancer Res. 73, 4830–4839 (2013)
Teves, S. S. & Henikoff, S. Transcription-generated torsional stress destabilizes nucleosomes. Nature Struct. Mol. Biol. 21, 88–94 (2014)
Veloso, A. et al. Genome-wide transcriptional effects of the anti-cancer agent camptothecin. PLoS ONE 8, e78190 (2013)
Abrahams, B. S. et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36 (2013)
Vaags, A. K. et al. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am. J. Hum. Genet. 90, 133–141 (2012)
van Daalen, E. et al. Social responsiveness scale-aided analysis of the clinical impact of copy number variations in autism. Neurogenetics 12, 315–323 (2011)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zylka, M., Philpot, B. & King, I. Zylka et al. reply. Nature 512, E2 (2014). https://doi.org/10.1038/nature13584
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13584
This article is cited by
-
Neurogenomics and the role of a large mutational target on rapid behavioral change
Biology Direct (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.