Abstract
The Palaeozoic form-taxon Lobopodia encompasses a diverse range of soft-bodied ‘legged worms’ known from exceptional fossil deposits1,2,3,4,5,6,7,8,9. Although lobopodians occupy a deep phylogenetic position within Panarthropoda, a shortage of derived characters obscures their evolutionary relationships with extant phyla (Onychophora, Tardigrada and Euarthropoda)2,3,5,10,11,12,13,14,15. Here we describe a complex feature in the terminal claws of the mid-Cambrian lobopodian Hallucigenia sparsa—their construction from a stack of constituent elements—and demonstrate that equivalent elements make up the jaws and claws of extant Onychophora. A cladistic analysis, informed by developmental data on panarthropod head segmentation, indicates that the stacked sclerite components in these two taxa are homologous—resolving hallucigeniid lobopodians as stem-group onychophorans. The results indicate a sister-group relationship between Tardigrada and Euarthropoda, adding palaeontological support to the neurological16,17 and musculoskeletal18,19 evidence uniting these disparate clades. These findings elucidate the evolutionary transformations that gave rise to the panarthropod phyla, and expound the lobopodian-like morphology of the ancestral panarthropod.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Whittington, H. B. The lobopod animal Aysheaia pedunculata Walcott, Middle Cambrian, Burgess Shale, British Columbia. Phil. Trans. R. Soc. Lond. B 284, 165–197 (1978)
Hou, X.-G. & Bergström, J. Cambrian lobopodians–ancestors of extant onychophorans? Zool. J. Linn. Soc. 114, 3–19 (1995)
Ramsköld, L. & Chen, J.-Y. in Arthropod Fossils and Phylogeny (ed. Edgecombe, G. D. ) 107–150 (Columbia Univ. Press, 1998)
Bergström, J. & Hou, X. Cambrian Onychophora or xenusians. Zool. Anz. 240, 237–245 (2001)
Ma, X., Edgecombe, G. D., Legg, D. A. & Hou, X. The morphology and phylogenetic position of the Cambrian lobopodian Diania cactiformis. J. Syst. Palaeontol. 12, 445–457 (2014)
Caron, J.-B., Smith, M. R. & Harvey, T. H. P. Beyond the Burgess Shale: Cambrian microfossils track the rise and fall of hallucigeniid lobopodians. Proc. R. Soc. B 280, 20131613 (2013)
Topper, T. P., Skovsted, C. B., Peel, J. S. & Harper, D. A. T. Moulting in the lobopodian Onychodictyon from the lower Cambrian of Greenland. Lethaia 46, 490–495 (2013)
Steiner, M., Hu, S.-X., Liu, J. & Keupp, H. A new species of Hallucigenia from the Cambrian Stage 4 Wulongqing Formation of Yunnan (South China) and the structure of sclerites in lobopodians. Bull. Geosci. 87, 107–124 (2012)
Ou, Q., Shu, D. & Mayer, G. Cambrian lobopodians and extant onychophorans provide new insights into early cephalization in Panarthropoda. Nature Commun. 3, 1261 (2012)
Budd, G. E. Tardigrades as ‘stem-group arthropods’: the evidence from the Cambrian fauna. Zool. Anz. 240, 265–279 (2001)
Liu, J. et al. An armoured Cambrian lobopodian from China with arthropod-like appendages. Nature 470, 526–530 (2011)
Legg, D. A. et al. Lobopodian phylogeny reanalysed. Nature 476, http://dx.doi.org/10.1038/nature10267 (10 August 2011)
Budd, G. E. The morphology of Opabinia regalis and the reconstruction of the arthropod stem-group. Lethaia 29, 1–14 (1996)
Wills, M. A., Briggs, D. E. G., Fortey, R. A., Wilkinson, M. & Sneath, P. H. A. in Arthropod Fossils and Phylogeny (ed. Edgecombe, G. D. ) 33–105 (Columbia Univ. Press, 1998)
Dewel, R. A., Budd, G. E., Castano, D. F. & Dewel, W. C. The organization of the subesophageal nervous system in tardigrades: insights into the evolution of the arthropod hypostome and tritocerebrum. Zool. Anz. 238, 191–203 (1999)
Mayer, G., Kauschke, S., Rüdiger, J. & Stevenson, P. A. Neural markers reveal a one-segmented head in tardigrades (water bears). PLoS ONE 8, e59090 (2013)
Mayer, G. et al. Selective neuronal staining in tardigrades and onychophorans provides insights into the evolution of segmental ganglia in panarthropods. BMC Evol. Biol. 13, 230 (2013)
Marchioro, T. et al. Somatic musculature of Tardigrada: phylogenetic signal and metameric patterns. Zool. J. Linn. Soc. 169, 580–603 (2013)
Schulze, C. & Schmidt-Rhaesa, A. Organisation of the musculature of Batillipes pennaki. Meiofauna Mar. 19, 195–207 (2011)
Budd, G. E. A palaeontological solution to the arthropod head problem. Nature 417, 271–275 (2002)
Daley, A. C., Budd, G. E., Caron, J.-B., Edgecombe, G. D. & Collins, D. H. The Burgess Shale anomalocaridid Hurdia and its significance for early euarthropod evolution. Science 323, 1597–1600 (2009)
Raup, D. M. Geometric analysis of shell coiling; general problems. J. Paleontol. 40, 1178–1190 (1966)
De Sena Oliveira, I. & Mayer, G. Apodemes associated with limbs support serial homology of claws and jaws in Onychophora (velvet worms). J. Morphol. 274, 1180–1190 (2013)
Robson, E. A. The cuticle of Peripatopsis moseleyi. Q. J. Microsc. Sci. s3–105, 281–299 (1964)
Olempska, E. Morphology and affinities of Eridostracina: Palaeozoic ostracods with moult retention. Hydrobiologia 688, 139–165 (2011)
Dzik, J. & Krumbiegel, G. The oldest ‘onychophoran’ Xenusion: a link connecting phyla? Lethaia 22, 169–181 (1989)
Engel, M. S., Davis, S. R. & Prokon, J. in Arthropod Biology and Evolution (eds Minelli, A., Boxshall, G. & Fusco, G. ) 269–298 (Springer, 2013)
Rota-Stabelli, O. et al. A congruent solution to arthropod phylogeny: phylogenomics, microRNAs and morphology support monophyletic Mandibulata. Proc. R. Soc. B 278, 298–306 (2011)
Campbell, L. I. et al. MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proc. Natl Acad. Sci. USA 108, 15920–15924 (2011)
Martin, C. & Mayer, G. Neuronal tracing of oral nerves in a velvet worm—implications for the evolution of the ecdysozoan brain. Front. Neuroanat. 8, 7 (2014)
Goloboff, P. A. Estimating character weights during tree search. Cladistics 9, 83–91 (1993)
Farris, J. S. A successive approximations approach to character weighting. Syst. Biol. 18, 374–385 (1969)
Mirande, J. M. Weighted parsimony phylogeny of the family Characidae (Teleostei: Characiformes). Cladistics 25, 574–613 (2009)
Nixon, K. C. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics 15, 407–414 (1999)
Goloboff, P. A. Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics 15, 415–428 (1999)
Goloboff, P. A., Farris, J. & Nixon, K. TNT: tree analysis using new technology. (http://www.lillo.org.ar/phylogeny/tnt/, 2003)
Goloboff, P. A., Farris, J. S. & Nixon, K. C. TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786 (2008)
Campiglia, S. & Lavallard, R. in Proc. 7th Int. Congr. Myriapodology (ed. Minelli, A. ) 461 (E. J. Brill, 1990)
Harvey, T. H. P., Ortega-Hernández, J., Lin, J.-P., Zhao, Y. & Butterfield, N. J. Burgess Shale-type microfossils from the middle Cambrian Kaili Formation, Guizhou Province, China. Acta Palaeontol. Pol. 57, 423–436 (2012)
Acknowledgements
The authors are supported by Research Fellowships at Clare College (M.R.S.) and Emmanuel College (J.O.-H.), University of Cambridge, UK. Thanks to J.-B. Caron and T. Harvey for images, access to material and discussions. D. Erwin, K. Hollis and P. Fenton facilitated access to museum specimens, and S. Whittaker assisted with electron microscopy. E. kanangrensis were collected from the Blue Mountains, New South Wales, with assistance from G. Budd and N. Tait and funding from an H.B. Whittington Research Grant (Paleontological Society). N. Butterfield and R. Janssen provided additional material. Parks Canada provided research and collection permits to Royal Ontario Museum teams led by D. Collins. The software TNT is funded by the Willi Hennig Society.
Author information
Authors and Affiliations
Contributions
M.R.S. conceived the project; dissected, described and interpreted specimens; and ran the phylogenetic analysis. J.O.-H. led the integration of developmental data into phylogenetic analysis and the interpretation of results. Both authors contributed equally to data analysis, discussion of results and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Claw measurements.
To reconstruct the relationship between the stacked constituent elements, a digital image of a sclerite (a) was duplicated, rotated and enlarged such that its outer sclerite precisely overlay the inner sclerite in the original image (b; the cyan image has been enlarged by 5% and rotated to match the inner sclerite in the yellow image). Repetition of this process demonstrates a logarithmic growth trajectory (c); a logarithmic spiral was fitted to this trajectory and its Raupian parameters22 calculated. This process was most accurate in the inner jaw elements, whose dentate margin provided multiple landmarks that allowed the precise fitting of subsequent images. Estimates were also obtained for the outer jaws and appendicular claws of Euperipatoides, and the claws and spines of H. sparsa. Hallucigenia spines demonstrated variability in Raup’s D because they are sometimes obliquely preserved, so the maximum value was taken as representative. The implied growth rate of 2.4 ± 2.7% in Euperipatoides sclerites (range 0–8%; measured from five inner and six outer jaw sclerites) cannot persist throughout the organism’s lifespan, since moulting consistently occurs every 2 weeks (ref. 38). Either moulting occurs less frequently in wild populations, the rate of growth varies during ontogeny or the constituent elements deform slightly during growth.
Extended Data Figure 2 Density of scaly ornament in a hallucigeniid spine with three constituent elements (Geological Survey of Canada 136958).
a, Complete spine, showing regions of enlargement; b, apex of spine showing tips of two internal elements; c, margins of two internal elements faintly visible (dotted lines); density of scales where three elements are superimposed is 0.050 scales per square micrometre; where two elements are superimposed it is 0.039 scales per square micrometre; for a single element it is 0.026 scales per square micrometre; slight deviation from a 3:2:1 ratio is attributed to decreased visibility of individual scales in occluded regions; d, up to five scales overlap; only two could overlap if scales were restricted to the front and back surfaces of a single element. Transmitted light images from multiple focal planes combined using CombineZM (A. Hadley). Scale bar, a, 100 µm; b, 40 µm; c, 10 µm; d, 5 µm.
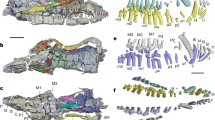
Extended Data Figure 3 Claws of Euperipatoides kanangrensis (Onychophora).
a, b, Secondary electron images of a single claw, separated into outermost element (a) and inner elements (b), each with ornamented basal region. c, d, Differential image contrast images of a single claw, separated into outermost element (c) and inner elements (d). Nomarski interference contrast accentuates the basal ornament. e–g, Single claw, separated into innermost element (e) and outer elements (f); pigmented foot tissue only associated with inner two elements; g, digital superposition of e and f showing original claw construction. h, Abnormal claw with blunt tip reflected in each constituent element. Transmitted light images from multiple focal planes combined using TuFuse (M. Lyons). Scale bar, 100 µm.
Extended Data Figure 4 Sclerite constitution in other taxa.
a, b, Single constituent element in claws of (a) Nephrotoma spp. (Tipulidae, Hexapoda, Euarthropoda) and (b) Eutardigrada (species indeterminate). Nomarski interference contrast. c–e, Small carbonaceous fossils with stacked constituent elements, interpreted as appendicular sclerites of total-group onychophorans (images courtesy of T. Harvey): c, d, from the basal mid-Cambrian (Stage 5) Kaili biota39; e, articulated pair from the mid- to late Cambrian Deadwood Formation, each claw comprising two constituent elements. f, Three appendicular sclerites (claws: arrowed) from a single appendage of Aysheaia pedunculata from the mid-Cambrian Burgess Shale (ROM 63052), each comprising a single element. Transmitted light images from multiple focal planes combined using TuFuse (M. Lyons) and CombineZM (A. Hadley). Scale bars, 100 µm.
Supplementary information
Supplementary Information
This file contains details of character coding for phylogenetic analysis, analytical methodology, and transformations implied by our most parsimonious tree. (PDF 1341 kb)
Supplementary Data
Zipped archive containing character matrix in Nexus and Excel formats, most parsimonious trees for all values of k in Nexus format (all individual trees, plus MPTs) and human-readable PDF format (MPTs only), and TNT script used to generate trees. (ZIP 515 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Smith, M., Ortega-Hernández, J. Hallucigenia’s onychophoran-like claws and the case for Tactopoda. Nature 514, 363–366 (2014). https://doi.org/10.1038/nature13576
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13576
This article is cited by
-
A multiscale approach reveals elaborate circulatory system and intermittent heartbeat in velvet worms (Onychophora)
Communications Biology (2023)
-
The velvet worm brain unveils homologies and evolutionary novelties across panarthropods
BMC Biology (2022)
-
Extensive loss of Wnt genes in Tardigrada
BMC Ecology and Evolution (2021)
-
Ancestral morphology of Ecdysozoa constrained by an early Cambrian stem group ecdysozoan
BMC Evolutionary Biology (2020)
-
Ecdysis in a stem-group euarthropod from the early Cambrian of China
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.