Abstract
N-methyl-d-aspartate (NMDA) receptors are Hebbian-like coincidence detectors, requiring binding of glycine and glutamate in combination with the relief of voltage-dependent magnesium block to open an ion conductive pore across the membrane bilayer. Despite the importance of the NMDA receptor in the development and function of the brain, a molecular structure of an intact receptor has remained elusive. Here we present X-ray crystal structures of the Xenopus laevis GluN1–GluN2B NMDA receptor with the allosteric inhibitor, Ro25-6981, partial agonists and the ion channel blocker, MK-801. Receptor subunits are arranged in a 1-2-1-2 fashion, demonstrating extensive interactions between the amino-terminal and ligand-binding domains. The transmembrane domains harbour a closed-blocked ion channel, a pyramidal central vestibule lined by residues implicated in binding ion channel blockers and magnesium, and a ∼twofold symmetric arrangement of ion channel pore loops. These structures provide new insights into the architecture, allosteric coupling and ion channel function of NMDA receptors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Traynelis, S. F. et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496 (2010)
Paoletti, P., Bellone, C. & Zhou, Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature Rev. Neurosci. 14, 383–400 (2013)
Bliss, T. V. P. & Collingridge, G. L. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361, 31–39 (1993)
Soto, D., Altafaj, X., Sindreu, C. & Bayes, A. Glutamate receptor mutations in psychiatric and neurodevelopmental disorders. Commun. Integr. Biol. 7, e27887 (2014)
Peery, H. E. et al. Anti-NMDA receptor encephalitis. The disorder, the diagnosis and the immunobiology. Autoimmun. Rev. 11, 863–872 (2012)
Keinänen, K. et al. A family of AMPA-selective glutamate receptors. Science 249, 556–560 (1990)
Bettler, B. et al. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron 5, 583–595 (1990)
Werner, P., Voigt, M., Keinänen, K., Wisden, W. & Seeburg, P. H. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature 351, 742–744 (1991)
Johnson, J. W. & Ascher, P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature 325, 529–531 (1987)
Mayer, M. L., Westbrook, G. L. & Guthrie, P. B. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309, 261–263 (1984)
Nowak, L., Bregestovski, P., Ascher, P., Herbet, A. & Prochiantz, A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature 307, 462–465 (1984)
Mayer, M. L. & Westbrook, G. L. Permeation and block of N-methyl-d-aspartic acid receptor channels by divalent cations in mouse cultured central neurones. J. Physiol. (Lond.) 394, 501–527 (1987)
Moriyoshi, K. et al. Molecular cloning and characterization of the rat NMDA receptor. Nature 354, 31–37 (1991)
Monyer, H. et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256, 1217–1221 (1992)
Tovar, K. R., McGinley, M. J. & Westbrook, G. L. Triheteromeric NMDA receptors at hippocampal synapses. J. Neurosci. 33, 9150–9160 (2013)
Hansen, K. B., Furukawa, H. & Traynelis, S. F. Control of assembly and function of glutamate receptors by the amino terminal domain. Mol. Pharmacol. 78, 535–549 (2010)
Kashiwagi, K. et al. Channel blockers acting at N-methyl-d-asparate receptors: differential effects of mutations in the vestibule and ion channel pore. Mol. Pharmacol. 61, 533–545 (2002)
Sun, Y. et al. Mechanism of glutamate receptor desensitization. Nature 417, 245–253 (2002)
Mayer, M. L. Emerging models of glutamate receptor ion channel structure and function. Structure 19, 1370–1380 (2011)
Pøhlsgaard, J., Frydenvang, K., Madsen, U. & Kastrup, J. S. Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology 60, 135–150 (2011)
Jin, R. et al. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 28, 1812–1823 (2009)
Kumar, J., Schuck, P., Jin, R. & Mayer, M. L. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nature Struct. Mol. Biol. 16, 631–638 (2009)
Karakas, E., Simorowski, N. & Furukawa, H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 28, 3910–3920 (2009)
Karakas, E., Simorowski, N. & Furukawa, H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2 NMDA receptors. Nature 475, 249–253 (2011)
Fischer, G. et al. Ro 25–6981, a highly potent and selective blocker of N-methyl-d-asparate receptors containing the NR2B subunit. Characterization in vitro. J. Pharmacol. Exp. Ther. 283, 1285–1292 (1997)
Watson, G. B. & Lanthorn, T. H. Pharmacological characteristics of cyclic homologues of glycine at the N-methyl-d-aspartate receptor-associated glycine site. Neuropharmacology 29, 727–730 (1990)
Allan, R. D. et al. Synthesis and activity of a potent N-methyl-D-aspartic acid agonist, trans-1-aminocyclobutane-1,3-dicarboxylic acid, and related phosphonic and carboxylic acids. J. Med. Chem. 33, 2905–2915 (1990)
Sobolevsky, A. I., Rosconi, M. P. & Gouaux, E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 (2009)
Salussolia, C. L., Prodromou, M. L., Borker, P. & Wollmuth, L. P. Arrangement of subunits in functional NMDA receptors. J. Neurosci. 31, 11295–11304 (2011)
Riou, M., Stroebel, D., Edwardson, J. M. & Paoletti, P. An alternating GluN1–2-1–2 subunit arrangement in mature NMDA receptors. PLoS ONE 7, e35134 (2012)
Lee, C. H. & Gouaux, E. Amino terminal domains of the NMDA receptor are organized as local heterodimers. PLoS ONE 6, e19180 (2011)
Furukawa, H., Singh, S., Mancusso, R. & Gouaux, E. Subunit arrangement and function in NMDA receptors. Nature 438, 185–192 (2005)
Mony, L., Kew, J. N., Gunthrope, M. J. & Paoletti, P. Allosteric modulators of NR2B-containing NMDA receptors: molecular mechanisms and therapeutic potential. Br. J. Pharmacol. 157, 1301–1317 (2009)
Reichling, D. B. & MacDermott, A. B. Lanthanum actions on excitatory amino acid-gated currents and voltage-gated calcium currents in rat dorsal horn neurons. J. Physiol. (Lond.) 441, 199–218 (1991)
Sherry, A. D., Newman, A. D. & Gutz, C. G. The activation of concavalin A by lanthanide ions. Biochemistry 14, 2191–2196 (1975)
Zhu, S., Stroebel, D., Yao, C. A., Taly, A. & Paoletti, P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nature Struct. Mol. Biol. 20, 477–485 (2013)
Weston, M. C., Schuck, P., Ghosal, A., Rosenmund, C. & Mayer, M. L. Conformational restriction blocks glutamate receptor desensitization. Nature Struct. Mol. Biol. 13, 1120–1127 (2006)
Gielen, M. et al. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron 57, 80–93 (2008)
Hansen, K. B., Ogden, K. K. & Traynelis, S. F. Subunit-selective allosteric inhbition of glycine binding to NMDA receptors. J. Neurosci. 32, 6197–6208 (2012)
Inanobe, A., Furukawa, H. & Gouaux, E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron 47, 71–84 (2005)
Erreger, K. et al. Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-d-aspartate glutamate receptors. Mol. Pharmacol. 72, 907–920 (2007)
Armstrong, N. & Gouaux, E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181 (2000)
Sugihara, H., Moriyoshi, K., Ishii, T., Masu, M. & Nakanishi, S. Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem. Biophys. Res. Commun. 185, 826–832 (1992)
Mony, L., Zhu, S., Carvalho, S. & Paoletti, P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. EMBO J. 30, 3134–3146 (2011)
Gielen, M., Siegler Retchless, B., Mony, L., Johnson, J. W. & Paoletti, P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 459, 703–707 (2009)
Yelshansky, M. V., Sobolevsky, A. I., Jatzke, C. & Wollmuth, L. P. Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J. Neurosci. 24, 4728–4736 (2004)
Erreger, K., Dravid, S. M., Banke, T. G., Wyllie, D. J. & Traynelis, S. F. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J. Physiol. (Lond.) 563, 345–358 (2005)
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998)
Burnashev, N. et al. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257, 1415–1419 (1992)
Kuner, T., Seeburg, P. H. & Guy, H. R. A common architecture for K+ channels and ionotropic glutamate receptors. Trends Neurosci. 26, 27–32 (2003)
Dukkipati, A., Park, H. H., Waghray, D., Fischer, S. & Garcia, K. C. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif. 62, 160–170 (2008)
Baconguis, I. & Gouaux, E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400–405 (2012)
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006)
Hattori, M., Hibbs, R. E. & Gouaux, E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 (2012)
Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl Acad. Sci. USA 99, 13419–13424 (2002)
Gourdon, P. et al. HiLiDe–Systematic approach to membrane protein crystallization in lipid and detergent. Cryst. Growth Des. 11, 2098–2106 (2011)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Hanson, M. A. et al. Crystal structure of a lipid-G protein-coupled receptor. Science 335, 851–855 (2012)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007)
Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D 59, 1131–1137 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Samsom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996)
DeLano, W. L. The PyMOL molecular graphic system (DeLano Scientific, San Carlos, California, USA, 2002)
Hart, H. E. & Greenwald, E. B. Scintillation proximity assay (SPA)—a new method of immunoassay. Direct and inhibition mode detection with human albumin and rabbit antihuman albumin. Mol. Immunol. 16, 265–267 (1979)
Acknowledgements
All members of the Gouaux laboratory are gratefully acknowledged for their support and assistance, especially L. Chen, K. Duerr and K. Wang. We thank L. Vaskalis for assistance with the figures, G. Westbrook and C. Jahr for comments on the manuscript, H. Owen for proofreading and I. Baconguis for making the animation. E.G. acknowledges the generous support of R. LaCroute, B. LaCroute and J. LaCroute. This work was also supported by an Oregon Brain Institute Graduate Student Fellowship (C.H.L.), the NIH (E.G.) and the Vollum Institute (E.G.). E.G. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
C.-H.L. contributed to all aspects of the research; W.L. carried out crystallographic analysis; J.C.M. carried out molecular biology, cell culture, electrophysiology and ligand binding experiments; A.G. performed molecular biology, cell culture, receptor purification and crystallization studies; J.D. and X.S. analysed the structures; E.G. directed the research; and all authors contributed to the preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
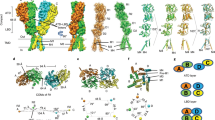
Extended Data Figure 1 Summary of Xenopus laevis NMDA crystallization constructs.
a, b, Cartoon representation of amino-terminal domain (ATD), ligand binding domain (LBD) and transmembrane domain (TMD) for GluN1 Δ2 (a) and GluN2B Δ2 (b) subunit constructs. Location of point mutations are highlighted in white circles. Location of deletions are highlighted with a yellow wedge. Mutated glycosylation sites are not shown and are listed in Extended Data Table 1. c, d, Select amino acid sequences of constructs used in these studies compared to wild-type sequence to highlight mutations in GluN1 (c) and GluN2B (d). Mutations are numbered and the purpose of each is detailed in Extended Data Table 1.
Extended Data Figure 2 Electrophysiology and western blot analysis of (GluN1 Δ)–(GluN2B Δ) receptor combinations.
a–c, Representative TEVC currents recorded for oocytes expressing GluN1 Δ4 and GluN2B Δ1 (a) or GluN2B Δ3 (b, c) receptors in response to agonist (100 μM glycine and 100 μM glutamate, bars, 20 s) or agonist plus 1 mM MgCl2 (indicated) after soaking oocytes in the absence (a, b) or presence (c) of 5 mM DTT. d, Western blot analysis of oocytes demonstrating spontaneously crosslinking cysteines (Lys216Cys) introduced at the GluN2B Δ3 intersubunit interface. Oocytes were soaked in the absence (left lanes) or presence of 5 mM DTT (right lanes) before processing for western analysis using an anti-GluN2B antibody. Filled and open triangles indicate positions of crosslinked and monomeric GluN2B, respectively. e, Graph of mean agonist-induced inward currents from four reduced oocytes expressing GluN1 Δ4 and GluN2B Δ3 in the absence (G/G, −25 ± −4 nA) or presence of 1 mM MgCl2 (G/G/Mg2+, 8 ± 5 nA). Error bars represent s.e.m. The P value is <0.001 for the paired t-test (asterisk). f, Representative TEVC currents recorded in response to agonist (100 μM glycine and 100 μM glutamate bars, 10 s) or agonist plus 1 mM MgCl2 for oocytes expressing constructs similar to the (GluN1 Δ2)–(GluN2B Δ2) receptor combination with the following exceptions: GluN1 subunit, Asp 656 (wild type), Gly636Arg and Lys741Asp; and GluN2B subunit, Glu 654 (wild type), Glu 655 (wild type), and Lys 216 (wild type). g, Binding constants for the (GluN1 Δ2)–(GluN2B Δ2) construct.
Extended Data Figure 3 2Fo-Fcelectron density maps of the GluN1–GluN2B NMDA structure.
a–d, The electron densities associated with the GluN1 ATD (chain A) contoured at 1.7 σ (a), the GluN1 LBD (chain A) contoured at 1.6 σ (b), the TMD of the entire tetrameric receptor contoured at 1.0 σ (c) and the TMD of a single GluN2B subunit (chain D), showing the pore loop, also contoured at 1.0 σ (d). Electron density maps and structures were derived from data set 1/structure 1 for panels a and b and from data set 2/structure 2 for panels c and d (see Extended Data Table 2).
Extended Data Figure 4 Analysis of spontaneous crosslinking of single cysteine point mutants introduced in the GluN2B ATD of the GluN1–GluN2B receptor complex.
a, Western blot analysis of single cysteine mutants in the α5 helix of the GluN2B subunit. Solubilized extracts of HEK293S GnTI− cells expressing a C-terminal GFP-StrepII tag GluN2B construct (GluN2B Δ1) containing mutants as indicated with untagged GluN1 (GluN1 Δ1) were analysed by western blot using an anti-GFP polyclonal antibody. The open and filled arrows correspond to monomeric and dimeric GluN2B bands, respectively. b, Coomassie stained SDS–PAGE analysis of spontaneous crosslinking of GluN2B K216C containing receptor. Left and right lanes illustrate samples with different concentrations of protein for GluN1–GluN2B and GluN1–GluN2B K216C receptors. The asterisk indicates GluN1 monomer and the open and filled arrows correspond to monomeric and dimeric GluN2B bands, respectively.
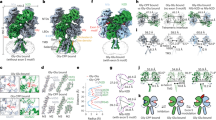
Extended Data Figure 5 Structural analyses and electron density maps of GluN1–GluN2B ATD heterodimer in the full-length NMDA structure.
a, Intersubunit distance between the indicated marker atoms and angle of domain closure in the soluble ATD structure (PDB 3QEM, left panel) or full-length ATD structure (right panel). b, Superposition of the full-length GluN1 (blue)–GluN2B (orange) ATD heterodimer onto the soluble heterodimer structure (PDB 3QEM, light grey) by aligning the indicated helices (green) in the R1 lobe of GluN2B. c, Fo-Fc omit electron density map for Ro25-6981 bound at the GluN1–GluN2B ATD heterodimer interface (chains A and B), contoured at 3 σ (data set 1/structure 1). d, Anomalous difference electron density of Tb3+ (blue mesh) near the R1–R2 hinge of a single GluN2B ATD (chain B, data set 3), contoured at 3.5 σ. e, Superposition of the LBD layer of the low resolution GluN1–GluN2B receptor (light blue, data set 4/structure 4) onto the LBD layer of the high resolution K216C receptor (magenta, data set 1/structure 1) illustrates the relative difference in ATD conformations between the two receptor structures (see Extended Data Table 2). Shown is the most open conformation of the ATDs derived from one of the two independent receptors in the asymmetric unit of data set 4/structure 4.
Extended Data Figure 6 LBD ligand electron densities and conformations.
a, b, Fo-Fc omit electron density maps for ACPC bound to GluN1 LBD (chain A) (a) and t-ACBD bound to GluN2B LBD (chain D) (b), contoured at 3 σ and 2.5 σ, respectively (data set 1/structure 1). c–e, Comparison of LBD in the full-length GluN1–GluN2B structure to isolated structures by aligning the D1 lobe. The angle of rotation relative to beta strand 10 is indicated for each. c, The ACPC-bound GluN1 LBD of the full-length structure (chain A, blue) is more open than the ACPC-bound isolated GluN1 LBD structure (PDB 1Y20, grey). d, The ACPC-bound GluN1 LBD of the full-length structure (chain A, blue) is more open than the glycine-bound isolated GluN1 LBD structure (PDB 2A5T, chain A, grey). e, The t-ACBD-bound GluN2B LBD of the full-length structure (chain D, orange) has a similar domain closure to the glutamate-bound isolated GluN2B LBD (PDB 2A5T, chain B, grey). f, GluN1–GluN2B LBD heterodimer (chains A and D) from the full-length receptor structure showing the separation of the D2 lobes, measured using the α-carbon atoms of residues Gly 664 and Gly 662, respectively. g, A similar measurement as in f using the equivalent residues in the context of the rat glycine/glutamate bound isolated GluN1–GluN2A LBDs (PDB 2A5T). h, The same measurement as in g, except in the GluN1 antagonist/Glu2A glutamate-bound conformation (PDB 4NF4). Structures shown in panels c–f were derived from data set 1/structure 1 and are similar in conformation to the related domains derived from data set 2/structure 2 (see Extended Data Table 2).
Extended Data Figure 7 Structural analyses of the transmembrane domain of NMDA receptor.
a, Alpha-carbon superposition of the M3 helices of the GluN1–GluN2B NMDA receptor (data set 2/structure 2) onto the corresponding M3 regions of GluA2 receptor (PDB 3KG2; grey). The r.m.s.d. is 1.89 Å for 144 aligned α-carbon atoms. The GluN1 subunits are blue and the GluN2B subunits are yellow. b, Amino acid sequence alignment of the NMDA receptor and the KcsA channel in the M2 and M3 regions using Promals3D (http://prodata.swmed.edu/promals3d/promals3d.php). c, Superposition of the four M2 helices of the NMDA receptor onto the corresponding four M2 regions of the KcsA channel (PDB 1K4C; residues 61–75). The r.m.s.d. is 1.86 Å. Only chains B and D of the NMDA GluN2B subunits are shown. d, Residual electron density in the central vestibule. Fo-Fc electron density in the central vestibule is shown for the GluN1–GluN2B receptor from data set 2/structure 2. For clarity, chain C is removed. e, Fo-Fc electron density map in the central vestibule derived from data set 1/structure 1. For clarity, chain B is removed. f, The same electron density map as shown in panel e except that the structure has been rotated by ∼90° around the pore axis and chain C of the GluN1 subunit has been removed for clarity. All maps are contoured at 2.8 σ.
Extended Data Figure 8 Comparison of LBD layers and LBD–TMD linkers between the NMDA receptor and the GluA2 receptor structures.
a, View from the extracellular side of the membrane of the connections between the TMD and LBD domains of the GluN1–GluN2B structure and of the GluA2 structure (PDB 3KG2), showing the relative rotation of GluA2 layer by ∼35°. The S2 segment resides within the LBD. The LBD-M3 linkers are highlighted. b, The LBD-M1 linkers are highlighted. c, The LBD-M4 linkers are highlighted. Shown in all panels are structures derived from data set 1/structure 1.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion. (PDF 108 kb)
Architecture of the full-length GluN1/GluN2B
The video shows the architecture of the heterotetrameric GluN1/GluN2B receptor in complex with ligands Ro25-6981, ACPC, and t-ACBD. GluN1 and GluN2B subunits are shown in both surface and cartoon representations colored blue and orange, respectively. Ligands Ro25-6981, ACPC, and t-ACBD are shown in sphere representation and colored green, red, and magenta, respectively. One ACPC and one t-ACBD molecule are included in the structure, bound to the GluN1 (chain A) and GluN2B (chain D) subunits, respectively. The video further presents the organization of domains ATD, LBD and TMD by rotating the molecule about the axes perpendicular and parallel to the membrane. Shown is Structure 2 derived from Data set 2 (see Extended Data Table 2). (MOV 22474 kb)
Rights and permissions
About this article
Cite this article
Lee, CH., Lü, W., Michel, J. et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511, 191–197 (2014). https://doi.org/10.1038/nature13548
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13548
This article is cited by
-
Adapting the endoplasmic reticulum proteostasis rescues epilepsy-associated NMDA receptor variants
Acta Pharmacologica Sinica (2024)
-
Disease-associated nonsense and frame-shift variants resulting in the truncation of the GluN2A or GluN2B C-terminal domain decrease NMDAR surface expression and reduce potentiating effects of neurosteroids
Cellular and Molecular Life Sciences (2024)
-
Two gates mediate NMDA receptor activity and are under subunit-specific regulation
Nature Communications (2023)
-
Unveiling the molecular basis of lobeline's allosteric regulation of NMDAR: insights from molecular modeling
Scientific Reports (2023)
-
Chemical shift assignments of calmodulin bound to the GluN1 C0 domain (residues 841–865) of the NMDA receptor
Biomolecular NMR Assignments (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.