Abstract
The surface of the cornea consists of a unique type of non-keratinized epithelial cells arranged in an orderly fashion, and this is essential for vision by maintaining transparency for light transmission. Cornea epithelial cells (CECs) undergo continuous renewal from limbal stem or progenitor cells (LSCs)1,2, and deficiency in LSCs or corneal epithelium—which turns cornea into a non-transparent, keratinized skin-like epithelium—causes corneal surface disease that leads to blindness in millions of people worldwide3. How LSCs are maintained and differentiated into corneal epithelium in healthy individuals and which key molecular events are defective in patients have been largely unknown. Here we report establishment of an in vitro feeder-cell-free LSC expansion and three-dimensional corneal differentiation protocol in which we found that the transcription factors p63 (tumour protein 63) and PAX6 (paired box protein PAX6) act together to specify LSCs, and WNT7A controls corneal epithelium differentiation through PAX6. Loss of WNT7A or PAX6 induces LSCs into skin-like epithelium, a critical defect tightly linked to common human corneal diseases. Notably, transduction of PAX6 in skin epithelial stem cells is sufficient to convert them to LSC-like cells, and upon transplantation onto eyes in a rabbit corneal injury model, these reprogrammed cells are able to replenish CECs and repair damaged corneal surface. These findings suggest a central role of the WNT7A–PAX6 axis in corneal epithelial cell fate determination, and point to a new strategy for treating corneal surface diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Davanger, M. & Evensen, A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature 229, 560–561 (1971)
Cotsarelis, G., Cheng, S. Z., Dong, G., Sun, T. T. & Lavker, R. M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell 57, 201–209 (1989)
Li, W. et al. Down-regulation of Pax6 is associated with abnormal differentiation of corneal epithelial cells in severe ocular surface diseases. J. Pathol. 214, 114–122 (2008)
Pellegrini, G. et al. p63 identifies keratinocyte stem cells. Proc. Natl Acad. Sci. USA 98, 3156–3161 (2001)
Mills, A. A. et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 (1999)
Yang, A. et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 (1999)
Koster, M. I., Kim, S., Mills, A. A., DeMayo, F. J. & Roop, D. R. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 18, 126–131 (2004)
Rama, P. et al. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 363, 147–155 (2010)
Blanpain, C. & Fuchs, E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nature Rev. Mol. Cell Biol. 10, 207–217 (2009)
Arwert, E. N., Hoste, E. & Watt, F. M. Epithelial stem cells, wound healing and cancer. Nature Rev. Cancer 12, 170–180 (2012)
Kopan, R. & Fuchs, E. A new look into an old problem: keratins as tools to investigate determination, morphogenesis, and differentiation in skin. Genes Dev. 3, 1–15 (1989)
Lauweryns, B., van den Oord, J. J. & Missotten, L. The transitional zone between limbus and peripheral cornea. An immunohistochemical study. Invest. Ophthalmol. Vis. Sci. 34, 1991–1999 (1993)
Eichner, R., Bonitz, P. & Sun, T. T. Classification of epidermal keratins according to their immunoreactivity, isoelectric point, and mode of expression. J. Cell Biol. 98, 1388–1396 (1984)
Schlötzer-Schrehardt, U. & Kruse, F. E. Identification and characterization of limbal stem cells. Exp. Eye Res. 81, 247–264 (2005)
Dorsky, R. I., Moon, R. T. & Raible, D. W. Control of neural crest cell fate by the Wnt signalling pathway. Nature 396, 370–373 (1998)
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011)
von Maltzahn, J., Bentzinger, C. F. & Rudnicki, M. A. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nature Cell Biol. 14, 186–191 (2012)
Yu, F.-X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012)
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001)
Fox-Walsh, K., Davis-Turak, J., Zhou, Y., Li, H. & Fu, X. D. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics 98, 266–271 (2011)
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998)
Xue, Y. et al. Direct conversion of fibroblasts to neurons by reprogramming PTB- regulated microRNA circuits. Cell 152, 82–95 (2013)
Acknowledgements
This study is supported in part by the 973 program (2013CB967504 and 2014CB964900), Project of Fundamental Research Funds (no.2012KF03), State Key laboratory of Ophthalmology, NIH (GM049369), KACST-UCSD Center of Excellence in Nanomedicine, NIH Director’s Transformative RO1 Program (R01 EY021374) and CIRM.
Author information
Authors and Affiliations
Contributions
H.O., X.-D.F., Yiz.L., and K.Z. designed study, interpreted data and wrote the manuscript. H.O., Y.X., Yin.L., X.Z., L.X., H.C., J.L., Mei.Z., Min.Z., Y.Y., H.L., G.L., E.Y., G.C., J.Z. and B.Y. performed the experiments. Y.L., W.J., J.L. and Yiz.L. obtained human samples. S.C., S.P., M.P. and L.Z. contributed to data analysis and interpretation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Keratin expression profiles and cell cultures, and three-dimensional differentiation of LSCs and SESCs.
a–f, Keratin expression profiles in human limbus, cornea and skin epidermis. a, b, Peripheral cornea–limbus junction and skin tissues showing K5+ (a) and K14 (b) expression in the basal cell layer of limbus and skin, and their absence in central corneal epithelium. c, d, Skin epidermis showing positive K1 (c) and K10 (d) expression and their absence in cornea and limbus. e, f, Peripheral cornea–limbus junction showing K19+ staining in limbus but not in central corneal epithelium and skin (e), and K3/12+ staining only in cornea and not in limbus and skin (f). g–j, Cultured LSCs with stem and progenitor cell characteristics, and SESC characteristics at passage 12 and validation of a three-dimensional differentiation system. g, Immunofluorescence staining of LSCs showing positive stem cell signals of p63 (a′) and Ki67 (b′) and negative differentiated CEC signals, K3/12 (c′), phase contrast photograph (d′). h, qPCR analysis showing K3/12 upregulation and K19 downregulation in CECs from a three-dimensional differentiation assay compared with LSCs. i, K1 and K10 upregulation in SECs from three-dimensional differentiation assay compared with SESCs (c), all n = 3, P < 0.01. j, Immunofluorescence staining of cultured SESCs showing positive p63 (a′) and negative signals for limbus stem cell marker, K19 (b′) and mature skin epithelium markers K1 and K10 (c′, d′). Scale bars, 100 µm. Data shown as means ± s.d.
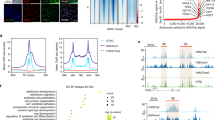
Extended Data Figure 2 Gene expression profiling and immunohistological analysis.
a–c, Genome-wide gene expression microarray of LSCs, CECs and SESCs. a, The top 100 significant genes in a comparison of LSCs and CECs to SESCs. b, Validation of the microarray data with qPCR analysis showing a strong correlation. c, qPCR analysis of WNT7A and PAX6 expression in LSCs and CECs compared to SESCs, all n = 3, P < 0.05. d, Expression of WNT7A and PAX6 in cornea and limbus of a one-year old human infant. H&E stain (a′), boxed area was shown in serial sections (b′–d′) with immunofluorescence staining of WNT7A (b′), PAX6 (c′) and K3/12 (d′). All scale bars, 100 µm. Data shown as means ± s.d.
Extended Data Figure 3 Appearance of skin epidermal markers with loss of corneal markers in human corneal diseases.
Appearance of skin epidermal markers p63, K5 and K10 with loss of corneal marker K3/12, PAX6 and WNT7A in cornea of patients with Stevens–Johnson syndrome (a, b), ocular pemphigoid (c), trauma injury (d) and alkaline burn (e). For all images, H&E staining was carried out on the lesion of corneal epithelial squamous metaplasia (a′). b′–f′, the same region of lesion in serial sections showing increased p63 (b′, d′) and K5 (c′, d′) and K10 (e′) in the suprabasal layer, no WNT7A (e′), K3/12 or PAX6 could be detected in the area (f′). Scale bars, 100 µm.
Extended Data Figure 4 The effect of WNT7A and FZD5 on PAX6 expression in LSCs.
a–c, The effect of WNT7A knockdown on PAX6 expression in LSCs. a, Phase contrast photographs showing effects of WNT7A and PAX6 knockdowns (shWNT7A and shPAX6) in LSCs and their three-dimensional differentiation spheres. b, qPCR analysis of gene expression changes of WNT7A and PAX6 in LSCs. WNT7A knockdown decreased PAX6 expression (n = 3, P < 0.01); no significant change in WNT7A expression in PAX6 knockdown. c, Validation of knockdown efficiency of WNT7A and PAX6 in LSCs by western blot analysis. d–f, WNT7A and FZD5 acted as the upstream regulators of PAX6 expression. d, Phase contrast photographs showing cell morphology of knockdown of FZD5 (shFZD5) in LSCs and three-dimensional differentiation spheres. e, Co-immunoprecipitation of WNT7A and FZD5 in LSCs. f, qPCR analysis of gene expression changes in corneal and skin epithelial markers in three-dimensional differentiated cells of LSCs with FZD5 knockdown (three-dimensional shFZD5 LSCs). FZD5 knockdown did not affect WNT7A expression; all others, n = 3, P < 0.05. Scale bars, 100 µm. Data shown as means ± s.d.
Extended Data Figure 5 The effect of PAX6 transduction in SESCs.
a, Phase contrast photographs of SESCs with PAX6 transduction (PAX6+) and three-dimensional differentiation spheres. b, Validation of K12 and PAX6 expression in three-dimensional differentiation spheres by western blotting analysis. c, Loss of skin-specific keratins, K1 and K10 in three-dimensional differentiation of SESCs with PAX6 transduction (three-dimensional PAX6+ SESCs). Scale bars, 100 µm.
Extended Data Figure 6 Quantitative information from RNA-seq data.
a, Statistical analysis of RNA-seq samples: raw reads, mapping reads and mapping rate of each sample are included. b, Pairwise comparisions of duplicated biological samples. c, The differences between SECs and three-dimensional PAX6+ SESCs, CECs and three-dimensional shPAX6 LSCs, all FDR < 0.001. a, qPCR analysis of PAX6 expression in rabbit SESCs with PAX6 transduction (rabbit (Rb) PAX6+ SESCs) or LSCs with PAX6 knockdown (Rb shPAX6 LSCs) (all n = 3, P < 0.05). We noticed some minor differences in the heatmap. These might result from some experimental variations, or it is possible that, although PAX6 expression is largely responsible for cell fate switch from SESCs to CECs at gene expression and functional levels (as demonstrated in this study), this single transcription factor may not be sufficient to create cells that are completely identical to CECs.
Extended Data Figure 7 Engineered expression of PAX6 and rabbit LSC deficiency model.
a–e, Quantification and culture of engineered expression of PAX6 in rabbit SESCs and PAX6 knockdown LSCs. a, qPCR analysis of PAX6 expression in rabbit SESCs with PAX6 transduction (Rb PAX6+ SESCs) or LSCs with PAX6 knockdown (Rb shPAX6 LSCs) (all n = 3, P < 0.05). b, Rabbit SESCs with positive staining of p63 and negative staining of PAX6. Left panel, phase contrast photograph. c, Top row, double immunofluorescence staining of PAX6 and p63 in rabbit SESCs with PAX6 transduction. Top left panel, phase contrast photograph. Bottom row, rabbit PAX6+ SESCs were further labelled with GFP for transplantation. d, Rabbit LSCs with positive staining of p63 and PAX6. Top left panel, phase contrast photograph. e, Culture of GFP-labelled rabbit LSCs with PAX6 knockdown. f, conjunctiva peritomy was performed and a circumferential strip of 2mm anterior limbal conjunctiva was removed (a′). Lamellar scleral and corneal dissection to completely remove LSCs and corneal epithelium along an anterior cornea stroma plane (b′–d′). Dissected cap is shown in (d′, arrows). The exposed cornea stroma bed was covered by human amniotic membrane (e′) and sutures (f′). (n = 3). Scale bars, 100 µm. Data shown as means ± s.d.
Extended Data Figure 8 Cornea epithelium regeneration and repair by transplanted GFP-labelled PAX6+ SESCs in a rabbit LSC deficiency model.
a, Time course of corneal epithelial defect repair. Fifteen days post transplantation, there was decreased cornea clarity with an entire corneal epithelial defect evidenced by fluorescein stain of cornea surface; 30 days post transplantation there was improved cornea clarity and reduced fluorescein staining of cornea epithelial defect; 45 and 90 days post transplantation there was restoration and maintenance of cornea clarity. b, c, Two other examples of regeneration and repair of rabbit corneal epithelial surface 90 days post transplantation with GFP-labelled PAX6+ SESCs showing complete repair and re-epithelization of corneal epithelial defects. Left panels, white light micrographs; middle panels, slit-lamp micrographs; right panels, fluorescein staining (note that bright spots on the corneal surface were due to camera light reflection, they were not epithelial defects) of corneal epithelium (n = 5). d, H&E stain of regeneration and repair of corneal epithelial surface in three separate rabbits 90 days post transplantation with GFP-labelled PAX6+ SESCs showing intact corneal epithelium histology.
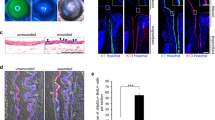
Extended Data Figure 9 Corneal epithelial regeneration by transplantation in a rabbit LSC deficiency model.
a, Time course of corneal epithelial regeneration and repair in a rabbit LSC deficiency model post transplantation with GFP-labelled PAX6+ SESCs. Top panels, 3 day post transplantation. left, light micrograph showing a hazy cornea ; right, GFP+ donor cells at limbal region (arrows). Bottom panels, 20 days post transplantation. Left, light micrograph showing a cornea with partial clarity; right, GFP+ donor co-located in transparent areas (arrows). Scale bars, 1 mm. We observed that only the transplanted cells from the limbal region could survive, proliferate and regenerate cornea surface epithelium, suggesting that limbus contained the stem cell niche favourable for stem cell survival and growth. b, Culture and re-isolation of reprogrammed donor GFP-labelled PAX6+ SESCs epithelial cells from the limbal region of a rabbit recipient eye 90 days post transplantation with GFP-labelled PAX6+ SESCs. Top panel, double immunofluorescence staining of PAX6 and GFP; bottom panel, double immunofluorescence staining of p63 and GFP in PAX6-transduced rabbit SESCs. Scale bars, 100 µm. c, Repair and recovery of a repeat cornea epithelium injury on a cornea transplanted with GFP-labelled PAX6+ SESCs. Top panels, we iatrogenically scraped and removed donor-derived corneal epithelial cells and made a large corneal surface epithelium defect (arrows) 3 months post initial transplantation of PAX6+ SESCs. Bottom panels, complete repair and recovery were observed within 72 h with healed epithelial defect (n = 3). Left panels, light micrographs; middle panels, slit-lamp micrographs; right panels, fluorescein staining.
Rights and permissions
About this article
Cite this article
Ouyang, H., Xue, Y., Lin, Y. et al. WNT7A and PAX6 define corneal epithelium homeostasis and pathogenesis. Nature 511, 358–361 (2014). https://doi.org/10.1038/nature13465
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13465
This article is cited by
-
RNA-Seq–based transcriptome analysis of corneal endothelial cells derived from patients with Fuchs endothelial corneal dystrophy
Scientific Reports (2023)
-
Single-cell atlas of keratoconus corneas revealed aberrant transcriptional signatures and implicated mechanical stretch as a trigger for keratoconus pathogenesis
Cell Discovery (2022)
-
Hsa-miR-143-3p inhibits Wnt-β-catenin and MAPK signaling in human corneal epithelial stem cells
Scientific Reports (2022)
-
Comprehensive 3D epigenomic maps define limbal stem/progenitor cell function and identity
Nature Communications (2022)
-
Multiplex viral tropism assay in complex cell populations with single-cell resolution
Gene Therapy (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.