Abstract
Srs2 helicase is known to dismantle nucleofilaments of Rad51 recombinase to prevent spurious recombination events1,2,3,4,5,6 and unwind trinucleotide sequences that are prone to hairpin formation7. Here we document a new, unexpected genome maintenance role of Srs2 in the suppression of mutations arising from mis-insertion of ribonucleoside monophosphates during DNA replication. In cells lacking RNase H2, Srs2 unwinds DNA from the 5′ side of a nick generated by DNA topoisomerase I8 at a ribonucleoside monophosphate residue. In addition, Srs2 interacts with and enhances the activity of the nuclease Exo1, to generate a DNA gap in preparation for repair. Srs2–Exo1 thus functions in a new pathway of nick processing-gap filling that mediates tolerance of ribonucleoside monophosphates in the genome. Our results have implications for understanding the basis of Aicardi–Goutières syndrome, which stems from inactivation of the human RNase H2 complex9.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Colavito, S. et al. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 37, 6754–6764 (2009)
Krejci, L. et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003)
Van Komen, S., Reddy, M. S., Krejci, L., Klein, H. & Sung, P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 278, 44331–44337 (2003)
Aboussekhra, A., Chanet, R., Adjiri, A. & Fabre, F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12, 3224–3234 (1992)
Chanet, R., Heude, M., Adjiri, A., Maloisel, L. & Fabre, F. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol. 16, 4782–4789 (1996)
Veaute, X. et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 (2003)
Bhattacharyya, S. & Lahue, R. S. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 24, 7324–7330 (2004)
Kim, N. et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332, 1561–1564 (2011)
Crow, Y. J. et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nature Genet. 38, 910–916 (2006)
Aguilera, A. & Klein, H. L. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119, 779–790 (1988)
Krejci, L. et al. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 279, 23193–23199 (2004)
Lisby, M., Rothstein, R. & Mortensen, U. H. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA 98, 8276–8282 (2001)
Nick McElhinny, S. A. et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl Acad. Sci. USA 107, 4949–4954 (2010)
Nick McElhinny, S. A. et al. Genome instability due to ribonucleotide incorporation into DNA. Nature Chem. Biol. 6, 774–781 (2010)
Sekiguchi, J. & Shuman, S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell 1, 89–97 (1997)
Chen, C., Umezu, K. & Kolodner, R. D. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2, 9–22 (1998)
Scheller, J., Schurer, A., Rudolph, C., Hettwer, S. & Kramer, W. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155, 1069–1081 (2000)
Genschel, J., Bazemore, L. R. & Modrich, P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 277, 13302–13311 (2002)
Cho, J. E., Kim, N., Li, Y. C. & Jinks-Robertson, S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair 12, 205–211 (2013)
Ghodgaonkar, M. M. et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell 50, 323–332 (2013)
Lujan, S. A., Williams, J. S., Clausen, A. R., Clark, A. B. & Kunkel, T. A. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell 50, 437–443 (2013)
Sparks, J. L. et al. RNase H2-initiated ribonucleotide excision repair. Mol. Cell 47, 980–986 (2012)
Spell, R. M. & Jinks-Robertson, S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 262, 3–12 (2004)
Adkins, N. L., Niu, H., Sung, P. & Peterson, C. L. Nucleosome dynamics regulates DNA processing. Nature Struct. Mol. Biol. 20, 836–842 (2013)
Seong, C., Colavito, S., Kwon, Y., Sung, P. & Krejci, L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J. Biol. Chem. 284, 24363–24371 (2009)
Fiorani, S., Mimun, G., Caleca, L., Piccini, D. & Pellicioli, A. Characterization of the activation domain of the Rad53 checkpoint kinase. Cell Cycle 7, 493–499 (2008)
Acknowledgements
We thank S. Jinks-Robertson and T. Kunkel for plasmids used in strain construction, R. Bermejo and M. Foiani for the genome sequencing of the original hpr4 strain, and G.-M. Li and S. Lee for providing hEXO1. This work was supported by National Institutes of Health grants RO1GM053738 (to H.L.K.), RO1ES007061 (to P.S.) and K99ES021441 (to H.N.).
Author information
Authors and Affiliations
Contributions
H.L.K. and P.S. conceived the study. C.J.P., H.N., P.S. and H.L.K. designed the experiments and analysed the data. C.J.P. and H.N. executed the experiments. C.J.P., H.N., P.S. and H.L.K. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
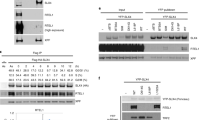
Extended Data Figure 1 Genetic interactions of the RNH201/202/203 genes and SRS2.
Lists of genes were obtained from the Saccharomyces Genome Database. Genes in grey are synthetically sick or synthetically lethal with either RNH201/RNH202/RNH203 or with SRS2. Genes in black are synthetically sick/lethal with both RNH201/RNH202/RNH203 and SRS2.
Extended Data Figure 2 Full-length srs2-KA shows the same double mutant phenotype with rnh202Δ as do srs2Δ and srs2-KA860.
a, Strains harbouring the helicase null K41A variant at the chromosomal SRS2 locus were constructed. b, Synthetic interaction of rnh202Δ and srs2-KA. c, Dinucleotide slippage rates; plotted are median rates with error bars representing 95% confidence intervals23 (n = 18).
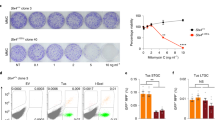
Extended Data Figure 3 Increased DNA damage load in the rnh202Δ srs2Δ double mutant.
a, Left, percentage cells with spontaneous Rad52–yellow fluorescent protein (YFP) foci formation, with at least ten fields of 50–200 cells counted per strain. Right, representative differential interference contrast and YFP images of a Rad52-YFP focus. b, Log-phase cells were analysed by flow cytometry to determine DNA content and data were quantified using FlowJo software. c, Percentage of large doublet cells in mid-log-phase cultures. Total number of cells and number of large doublet cells were determined for each strain by examining at least ten fields of 50–200 cells per strain. d, Wild-type and rnh202Δ srs2Δ double mutant cells were grown in YPD media, washed and treated for 1 h with 100 mM hydroxyurea. Lysates were extracted and subjected to immunoblotting with an anti-Rad53 antibody26. The higher species represents phosphorylated Rad53. Plotted in a and c are the mean values ± s.d. (n = 3).
Extended Data Figure 4 Mutation spectra at the CAN1 locus for the rnh202Δ single and rnh202Δ srs2Δ double mutant cells.
Independent mutations in the rnh202Δ (blue) and rnh202Δ srs2Δ (red) Canr mutant strains. Base substitutions are written above the wild-type sequence, deletions are represented by ‘ = ’ and insertions are represented by  .
.
Extended Data Figure 5 Unwinding of nicked duplex DNA by Srs2.
Intact duplex DNA (a) and nicked duplex DNA labelled at either the (b) 5′ or (c) 3′ end of the nicked strand were incubated with Srs2 for 10 min at 30 °C with or without ATP. Reactions were run in a polyacrylamide gel and analysed by phosphorimaging analysis. The results are quantified in d. Plotted are the mean values ± s.d. (n = 3).
Extended Data Figure 6 Topo I-catalysed DNA cleavage at the rUMP site.
a, Duplex DNA with or without an embedded rUMP residue (5 nM) was incubated with either E. coli or calf thymus Topo I for 30 min at 37 °C. b, Reactions were analysed by electrophoresis in a polyacrylamide gel followed by phosphorimaging analysis. c, Schematic for the construction of the DNA substrate that harbours a unique nick induced by calf thymus Topo I at a rUMP residue.
Extended Data Figure 7 Mph1 does not act at the Topo-I-induced nick.
a, DNA substrate (5 nM) that harbours a Topo-I-induced nick and radiolabelled on the 3′ side was incubated with Srs2 or Mph1 (20 or 40 nM) in the presence or absence of ATP at 30 °C for 10 min. The reactions were analysed as before. b, (AG)4 dinucleotide slippage rates; plotted are median rates, with error bars representing 95% confidence intervals23 (n = 18). c, Exo1 shows 5′ endo- and exonuclease activities on the 5′ Flap and the nicked duplex DNA, respectively. Radiolabelled 5′ Flap without or with a gap region, nicked duplex DNA, or oligonucleotide dT (5 nM each) was incubated with Exo1 for 10 min at 30 °C. The reactions were analysed as before and the products indicated by the arrows.
Extended Data Figure 8 Stimulation of Exo1-catalysed digestion of a 5′ Flap and a Topo-I-induced nick by Srs2.
a, Radiolabelled 5′ Flap substrate (5 nM) was incubated with Exo1 (0.1 nM) in the absence or presence of Mph1 for 10 min at 30 °C. b, Radiolabelled 5′ Flap substrate (5 nM) was incubated with hEXO1 (4 nM) in the absence or presence of Srs2 (0.4, 2 and 10 nM) for 10 min at 37 °C. c, Radiolabelled DNA substrate harbouring a Topo-I-induced nick (5 nM) was incubated with Exo1 (0.1 nM) in the absence or presence of Srs2 or Mph1 for 10 min at 30 °C. d, Radiolabelled substrate harbouring a Topo-I-induced nick (5 nM) was incubated with increasing amounts of hEXO1 (0.25, 0.5 and 1 nM) in the absence or presence of Srs2 (10 nM) for 10 min at 37 °C. The reactions in a–d were analysed as before. Plotted in b and d are the mean values ± s.d. (n = 3).
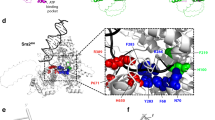
Extended Data Figure 9 Model of Srs2–Exo1-mediated processing of a DNA nick induced by Top1 at an embedded ribonucleotide.
RNase H2 normally removes misincorporated rNMPs from DNA (not shown). In the absence of RNase H2, topoisomerase 1 can cleave at the embedded ribonucleotide, leaving 3′ cyclic phosphate and 5′ OH ends that cannot be ligated. Repair starts with unwinding of DNA from the 5′ OH side by Srs2, followed by DNA digestion by Exo1 through its exonuclease or 5′ Flap endonuclease activity to result in a DNA gap, to be filled by a DNA polymerase. Our results show that Srs2 enhances the nuclease activities of Exo1. In the absence of Srs2, re-ligation of ends processed by unknown nuclease activities leads to a deletion mutation.
Rights and permissions
About this article
Cite this article
Potenski, C., Niu, H., Sung, P. et al. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature 511, 251–254 (2014). https://doi.org/10.1038/nature13292
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13292
This article is cited by
-
Signatures of TOP1 transcription-associated mutagenesis in cancer and germline
Nature (2022)
-
RNases H1 and H2: guardians of the stability of the nuclear genome when supply of dNTPs is limiting for DNA synthesis
Current Genetics (2020)
-
Apn2 resolves blocked 3′ ends and suppresses Top1-induced mutagenesis at genomic rNMP sites
Nature Structural & Molecular Biology (2019)
-
Resolution of a complex crisis at DNA 3′ termini
Nature Structural & Molecular Biology (2019)
-
The etiology of uracil residues in the Saccharomyces cerevisiae genomic DNA
Current Genetics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.