Abstract
The large spectrum of limb morphologies reflects the wide evolutionary diversification of the basic pentadactyl pattern in tetrapods. In even-toed ungulates (artiodactyls, including cattle), limbs are adapted for running as a consequence of progressive reduction of their distal skeleton to symmetrical and elongated middle digits with hoofed phalanges. Here we analyse bovine embryos to establish that polarized gene expression is progressively lost during limb development in comparison to the mouse. Notably, the transcriptional upregulation of the Ptch1 gene, which encodes a Sonic hedgehog (SHH) receptor, is disrupted specifically in the bovine limb bud mesenchyme. This is due to evolutionary alteration of a Ptch1 cis-regulatory module, which no longer responds to graded SHH signalling during bovine handplate development. Our study provides a molecular explanation for the loss of digit asymmetry in bovine limb buds and suggests that modifications affecting the Ptch1 cis-regulatory landscape have contributed to evolutionary diversification of artiodactyl limbs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Gene Expression Omnibus
Data deposits
All 4C-seq data sets are deposited in the Gene Expression Omnibus repository under accession number GSE52988. The GLI3 ChIP-seq data relevant to this study are deposited in the Gene Expression Omnibus repository under accession number GSE52939. All the raw data for sequencing of the Ptch1 locus are deposited in the European Nucleotide Archive (study accession number PRJEB5056).
References
Polly, P. D. Limbs in mammalian evolution. In Fins into Limbs ( Hall, B. K. ed.) 245–268 (Univ. Chicago Press, 2007)
Prothero D. R., Foss S. E., eds. The Evolution of Artiodactyls (Johns Hopkins Univ. Press, 2007)
Spaulding, M., O’Leary, M. A. & Gatesy, J. Relationships of Cetacea (Artiodactyla) among mammals: increased taxon sampling alters interpretations of key fossils and character evolution. PLoS ONE 4, e7062 (2009)
Zeder, M. A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl Acad. Sci. USA 105, 11597–11604 (2008)
Budras K. D., Habel R. E., eds. Bovine Anatomy (Schlütersche, 2011)
de Bakker, M. A. et al. Digit loss in archosaur evolution and the interplay between selection and constraints. Nature 500, 445–448 (2013)
Cooper, K. L. et al. Patterning and post-patterning modes of evolutionary digit loss in mammals. Nature (in the press)
Sears, K. E. et al. Developmental basis of mammalian digit reduction: a case study in pigs. Evol. Dev. 13, 533–541 (2011)
Benazet, J. D. et al. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science 323, 1050–1053 (2009)
Lopez-Rios, J. et al. GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev. Cell 22, 837–848 (2012)
Zeller, R., Lopez-Rios, J. & Zuniga, A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nature Rev. Genet. 10, 845–858 (2009)
Carroll, S. B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 (2008)
Galli, A. et al. Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 6, e1000901 (2010)
te Welscher, P., Fernandez-Teran, M., Ros, M. A. & Zeller, R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 16, 421–426 (2002)
Marigo, V., Johnson, R. L., Vortkamp, A. & Tabin, C. J. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev. Biol. 180, 273–283 (1996)
Zhu, J. et al. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell 14, 624–632 (2008)
Vokes, S. A., Ji, H., Wong, W. H. & McMahon, A. P. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 (2008)
Woltering, J. M. & Duboule, D. The origin of digits: expression patterns versus regulatory mechanisms. Dev. Cell 18, 526–532 (2010)
Lewandoski, M., Sun, X. & Martin, G. R. Fgf8 signalling from the AER is essential for normal limb development. Nature Genet. 26, 460–463 (2000)
Mariani, F. V., Ahn, C. P. & Martin, G. R. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature 453, 401–405 (2008)
Sanz-Ezquerro, J. J. & Tickle, C. Fgf signaling controls the number of phalanges and tip formation in developing digits. Curr. Biol. 13, 1830–1836 (2003)
Chiang, C. et al. Manifestation of the limb prepattern: limb development in the absence of Sonic Hedgehog Function. Dev. Biol. 236, 421–435 (2001)
Lettice, L. A. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 (2003)
Sagai, T. et al. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh). Mamm. Genome 15, 23–34 (2004)
Sagai, T., Hosoya, M., Mizushina, Y., Tamura, M. & Shiroishi, T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132, 797–803 (2005)
Shapiro, M. D., Hanken, J. & Rosenthal, N. Developmental basis of evolutionary digit loss in the Australian lizard Hemiergis. J. Exp. Zool. B 297, 48–56 (2003)
Harfe, B. D. et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004)
Gritli-Linde, A., Lewis, P., McMahon, A. P. & Linde, A. The whereabouts of a morphogen: direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Dev. Biol. 236, 364–386 (2001)
Briscoe, J., Chen, Y., Jessell, T. M. & Struhl, G. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol. Cell 7, 1279–1291 (2001)
Chen, Y. & Struhl, G. Dual roles for patched in sequestering and transducing hedgehog. Cell 87, 553–563 (1996)
Marigo, V., Davey, R. A., Zuo, Y., Cunningham, J. M. & Tabin, C. J. Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176–179 (1996)
Bouldin, C. M., Gritli-Linde, A., Ahn, S. & Harfe, B. D. Shh pathway activation is present and required within the vertebrate limb bud apical ectodermal ridge for normal autopod patterning. Proc. Natl Acad. Sci. USA 107, 5489–5494 (2010)
Vokes, S. A. et al. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977–1989 (2007)
Cotney, J. et al. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 22, 1069–1080 (2012)
Friedli, M. et al. A systematic enhancer screen using lentivector transgenesis identifies conserved and non-conserved functional elements at the Olig1 and Olig2 locus. PLoS ONE 5, e15741 (2010)
Goodrich, L. V., Milenkovic, L., Higgins, K. M. & Scott, M. P. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997)
van de Werken, H. J. et al. 4C Technology: Protocols and Data Analysis. Methods Enzymol. 513, 89–112 (2012)
van de Werken, H. J. et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nature Methods 9, 969–972 (2012)
Butterfield, N. C. et al. Patched 1 is a crucial determinant of asymmetry and digit number in the vertebrate limb. Development 136, 3515–3524 (2009)
Chan, Y. F. et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (2010)
Gompel, N., Prud’homme, B., Wittkopp, P. J., Kassner, V. A. & Carroll, S. B. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487 (2005)
McGregor, A. P. et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448, 587–590 (2007)
Cretekos, C. J. et al. Regulatory divergence modifies limb length between mammals. Genes Dev. 22, 141–151 (2008)
Davidson, E. H. & Erwin, D. H. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 (2006)
Peter, I. S. & Davidson, E. H. Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985 (2011)
Stern, D. L. & Frankel, N. The structure and evolution of cis-regulatory regions: the shavenbaby story. Phil. Trans. R. Soc. B 368, 1632 (2013)
Stern, D. L. The genetic causes of convergent evolution. Nature Rev. Genet. 14, 751–764 (2013)
Clifford, A. B. The evolution of the unguligrade manus in artiodactyls. J. Vertebr. Paleontol. 30, 1827–1839 (2010)
Ross, D. G., Bowles, J., Hope, M., Lehnert, S. & Koopman, P. Profiles of gonadal gene expression in the developing bovine embryo. Sex Dev. 3, 273–283 (2009)
Ellis, T. et al. Patched 1 conditional null allele in mice. Genesis 36, 158–161 (2003)
Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002)
van den Brandt, J., Wang, D., Kwon, S. H., Heinkelein, M. & Reichardt, H. M. Lentivirally generated eGFP-transgenic rats allow efficient cell tracking in vivo. Genesis 39, 94–99 (2004)
Quintana, L. & Sharpe, J. Preparation of mouse embryos for optical projection tomography imaging. CSH Protocols 2011, 664–669 (2011)
Koyama, E. et al. Development of stratum intermedium and its role as a Sonic hedgehog-signaling structure during odontogenesis. Dev. Dyn. 222, 178–191 (2001)
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Rosenbloom, K. R. et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41, D56–D63 (2013)
Eggen, A. et al. Construction and characterization of a bovine BAC library with four genome-equivalent coverage. Genet. Sel. Evol. 33, 543–548 (2001)
Schibler, L. et al. Construction and extensive characterization of a goat bacterial artificial chromosome library with threefold genome coverage. Mamm. Genome 9, 119–124 (1998)
Vaiman, D. et al. Construction and characterization of a sheep BAC library of three genome equivalents. Mamm. Genome 10, 585–587 (1999)
Rogel-Gaillard, C., Bourgeaux, N., Billault, A., Vaiman, M. & Chardon, P. Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet. Cell Genet. 85, 205–211 (1999)
Simpson, J. T. et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 (2009)
Acknowledgements
We thank K. Cooper and C. Tabin for sharing their results before publication and providing camel genomic DNA. Dolphin samples were received from C. Frere and D. Duffield. Pigmy hippopotamus mouth swabs for DNA extraction were provided by S. Furrer and B. Zimmerman from the Zoo of Zurich, whereas all other artiodactyl blood samples were donated from available frozen stocks by S. and M. Hoby from the Zoo of Basel. B. Wainwright provided the Ptch1 conditional mouse line. We are grateful to E. Terszowska and A. Offinger for mouse care; V. Metis and M. Rondon for assistance in colony maintenance and collection of Prx1-Cre Ptch1Δc/Δc mouse embryos; and N. Dumesnil for assistance in collecting bovine embryos. All lentivector-mediated transgenic mouse embryos were produced by the EPFL transgenic platform, whereas conventional mouse transgenic embryos were produced commercially by Cyagen Biosciences Inc. We thank S. Beck-Cormier, A. Gritli-Linde, D. Haag-Wackernagel, Y. Lallemand, P. Zimmermann, G. Nusspaumer and A. Zuniga for technical advice and input and M. Bertaud, I. Ginez, D. Jardet and M. Moroldo for technical assistance. We are grateful to V. Taylor and members of our research groups for critical discussions and input on the manuscript. This research was supported by SNF grants 31003A_130803/146248 and the University of Basel (to R.Z.), EU reintegration grant PERG-GA-2009-246576 (to J.L.-R.), SystemsX.ch iPhD Grant 20101078 (to D.I. and R.Z.), INRA (to A.D.), ANR grant No. 06-MRAR-027-01 (to B.R.), the EPFL and ERC grant SystemsHox.ch (to D.D.), NIH grant no. NS 033642 (to A.P.M.) and Australian National Health and Medical Research Council grant no. 569713 (to C.W.).
Author information
Authors and Affiliations
Contributions
J.L.-R., A.D. and R.Z. conceived the project and wrote the manuscript; J.L.-R. and A.D. performed most of the mouse and bovine experimental studies. D.S. and E.U. performed mouse and bovine experiments under supervision of J.L.-R. and R.Z.; J.L-.R. and D.S. isolated, sequenced and analysed the ∼9-kb LRM from different species. G.A. performed the lentivector studies under supervision of D.D. J.L. generated the Gli33XFlag mouse strain and K.A.P. performed the GLI3 ChIP-seq analysis under supervision of A.P.M. P.G. performed the in silico simulations under supervision of D.I. S.F. participated in the design of bovine ISH probes and together with A.D., S.B. and Y.G. produced and collected the bovine embryos for the study. M.M.-G. did the skeletal analysis of bovine foetuses using CT scanning. A.D.C. and C.W. provided all mouse embryos lacking Ptch1 in the limb bud mesenchyme. C.B. performed all deep sequencing of the Ptch1 loci from different species and as part of the 4C analysis. R.I. assisted in high-throughput data analysis. C.K. and S.R. participated in in silico assembly and analysis of the Ptch1 loci from different species. B.R. was key to conceiving and initiating this project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Equivalent stages of mouse and bovine fore- and hindlimb development.
a, Table listing the developmentally equivalent bovine fore- and hindlimb bud stages in comparison to mouse forelimb buds. CRL, approximate crown-rump length as previously defined49. b, False-coloured CT scan of a bovine fetus (at 5 months of gestation) to detect the ossified skeletal bones. Left limbs were coloured blue and right limbs orange. Radius and ulna form as two distinct skeletal elements, while the metacarpals/metatarsals fuse during ossification to give rise to the cannon bone, a convergent trait in many artiodactyls2. Lower panels illustrate the similarities of the forelimb (FL) and hindlimb (HL) autopod skeletal bones. Arrowheads indicate the plane of fusion of the metacarpal/metatarsal bones. Two independent fetuses were analysed. s, scapula; h, humerus; r, radius; u, ulna; c*, region corresponding to the not yet ossified carpus; cb, cannon bone; p, phalanges. III, IV, digits.
Extended Data Figure 2 Apoptosis in bovine forelimb buds during digit formation and gene expression during the onset of limb bud development.
a, The SOX9 transcriptional regulator (red immunofluorescence) marks all chondrogenic progenitors of the forming digit primordia in the bovine forelimb autopod. Apoptotic cells (TUNEL; green fluorescence) are only detected in the interdigit region that is being eliminated at these developmental stages (arrowhead in top panel). No apoptosis is detected in the digit-forming territories, except in the forming joints (arrowhead in bottom panel). Sections were counterstained with Hoechst-33258 to label cell nuclei. b, During initiation of limb bud outgrowth, the complementary expression of Gli3 and Hand2 marks the establishment of anterior–posterior asymmetry and is required to activate Shh expression. Mesenchymal cells responding to SHH signalling upregulate the expression of the SHH receptor Ptch1. Msx2 is a transcriptional sensor of BMP activity and its distribution marks territories of active BMP signalling. The expression of these genes is comparable in early mouse and bovine forelimb buds. In all Extended Data figures, limb buds are oriented with anterior to the top. Per marker and species, the distribution was reproduced in at least n = 3 independent limb bud samples in independent experiments. Scale bar, 0.25 mm.
Extended Data Figure 3 Validation of the whole-mount in situ hybridization probes and techniques in bovine embryos.
a, b, OPT analysis of Hoxd13 transcripts in the mesenchyme of mouse (a) and bovine (b) forelimbs. White arrowheads indicate the planes of the virtual OPT sections shown in the right panels. This analysis establishes that mesenchymal transcripts are equally well detected in mouse and bovine limb buds. Note the distal shift and more symmetric expression of Hoxd13 transcripts in bovine limb buds. c, Bovine and mouse embryos hybridized with the riboprobes for Shh, Ptch1, Gli1 and Hoxd13. No apparent changes in their expression in tissues other than limb buds were detected by comparing bovine and mouse embryos. Per gene and species, the transcript distribution was reproduced in at least n = 3 independent samples in independent experiments. Scale bar, 0.25 mm.
Extended Data Figure 4 Ptch1 and Ptch2 expression in limb buds.
a, OPT analysis of Ptch1 expression in mouse and bovine limb buds. White arrowheads indicate the levels of optical sections shown in the middle panels. Right panels show enlargements of the posterior part. b, Ptch2 expression in mouse and bovine limb buds. White arrowheads indicate the levels of optical sections shown in the right panels. Per gene and species, the transcript distribution was reproduced in at least n = 3 independent limb bud samples in independent experiments. Scale bars, 0.25 mm.
Extended Data Figure 5 Mathematical simulation of the mesenchymal response to SHH signal transduction with intact (mouse) and absent (bovine) Ptch1 upregulation in limb buds.
a, Simulations in wild-type mouse limb buds using the following minimal regulatory network (P. Germann and D. Iber, unpublished data). In the absence of Shh, Ptch1 promotes the formation of Gli repressor (predominantly Gli3R, indicated in blue) and antagonizes (in red) the formation of Gli activators (Gli3A, but also representing contributions by Gli2). Ptch1 is also genetically required for Gli3 expression in limb buds. Binding of Shh to Ptch1 (PS) relieves the tonic inhibition of Smo by Ptch1 (not shown), which promotes activator and inhibits repressor formation. Both Gli3A and Gli3R isoforms impact on Ptch1 and Gli1 expression, the expression kinetics of which were simulated (middle and right panels). For details of network construction and all references see Supplementary Note. In wild-type mouse limb buds, Ptch1 and Gli1 are upregulated in the posterior mesenchyme. b, Simulation of Ptch1 and Gli1 expression in bovine limb buds. The impact of Gli3A and Gli3R on Ptch1 transcription was removed from the network and the rate of Ptch1 production fixed at constant low levels. Simulations using this altered network result in progressive anterior expansion of Gli1 expression. c, Simulations of hypothetical bovine limb buds with an intact network results in posterior upregulation of both Ptch1 and Gli1 expression. d, Increasing Shh diffusion rates by tenfold (green dot in scheme) in an otherwise intact network leads to upregulation and anterior expansion of both Ptch1 and Gli1 expression, that is, fails to reproduce the real gene expression patterns. Broken lines in all panels indicate the simulated Ptch1 and Gli1 transcriptional activities at the indicated time points and the grey shaded area bordered by a solid line corresponds to the overall transcript levels. Red dotted line marks the position of peak expression. Gene symbols correspond to the computational variables used for simulations. A, anterior; P, posterior; h, hours; d, days. For detailed description of all simulations see Supplementary Note.
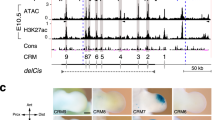
Extended Data Figure 6 Functional analysis of the conserved mouse and bovine core regions A and B using lentiviral transgenesis.
a, A variable and artiodactyl-specific microsatellite expansion (CA/CG2-27, red shading) in region A is located just upstream of exon 17 (Fig. 5a). b, Expression of a lacZ reporter inserted into the Ptch1 locus. c, d, Expression of lentivector ßlacZ reporter constructs controlled by the mouse and bovine conserved regions A and B (Fig. 5a) in transgenic mouse embryos (mouse: n = 12 of 41; bovine: n = 12 of 20 embryos). FL, forelimb. Scale bar, 0.25 mm.
Extended Data Figure 7 Functional analysis of the mouse and bovine LRM.
a, Expression of the mouse and bovine LRM in mouse hindlimb buds (E11.5). b, Transgenic mouse embryos collected at different stages reveal the temporal kinetics of activation and upregulation of the mouse LRM-ßlacZ transgenic reporter in forelimb buds (n = 3 independent embryos per stage). c, Forelimb buds of n = 6 independent transgenic embryos expressing the lacZ reporter under control of the bovine LRM region. The variable expression is in all cases restricted to the core mesenchyme (compare to Fig. 5d). Scale bar, 0.25 mm.
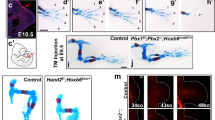
Extended Data Figure 8 Prx1-Cre-mediated conditional inactivation of Ptch1 in the mouse limb mesenchyme phenocopies the loss of asymmetry and alterations in digit primordia similar to bovine handplates.
a, b, Detection of Hoxd13 (a) and Sox9 (b) transcripts in mouse (left), bovine (middle) and Ptch1Δc/Δc (right panels) forelimb buds. Per gene, species and mouse genotype, the transcript distribution was reproduced in at least n = 3 independent limb bud samples in independent experiments. Vestigial digit primordia are indicated in red. Asterisks indicate digit primordia with uncertain identities. Scale bars, 0.25 mm.
Supplementary information
Supplementary Information
This file contains a Supplementary Note related to mathematical simulations and Supplementary Tables 1-3 related to the online methods. (PDF 426 kb)
Rights and permissions
About this article
Cite this article
Lopez-Rios, J., Duchesne, A., Speziale, D. et al. Attenuated sensing of SHH by Ptch1 underlies evolution of bovine limbs. Nature 511, 46–51 (2014). https://doi.org/10.1038/nature13289
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13289
This article is cited by
-
The little skate genome and the evolutionary emergence of wing-like fins
Nature (2023)
-
Evolutionary genetics of flipper forelimb and hindlimb loss from limb development-related genes in cetaceans
BMC Genomics (2022)
-
GLI transcriptional repression is inert prior to Hedgehog pathway activation
Nature Communications (2022)
-
Gene expression changes during the evolution of the tetrapod limb
Biologia Futura (2022)
-
Spatial regulation by multiple Gremlin1 enhancers provides digit development with cis-regulatory robustness and evolutionary plasticity
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.