Abstract
Insulin constitutes a principal evolutionarily conserved hormonal axis for maintaining glucose homeostasis1,2,3; dysregulation of this axis causes diabetes2,4. PGC-1α (peroxisome-proliferator-activated receptor-γ coactivator-1α) links insulin signalling to the expression of glucose and lipid metabolic genes5,6,7. The histone acetyltransferase GCN5 (general control non-repressed protein 5) acetylates PGC-1α and suppresses its transcriptional activity, whereas sirtuin 1 deacetylates and activates PGC-1α8,9. Although insulin is a mitogenic signal in proliferative cells10,11, whether components of the cell cycle machinery contribute to its metabolic action is poorly understood. Here we report that in mice insulin activates cyclin D1–cyclin-dependent kinase 4 (Cdk4), which, in turn, increases GCN5 acetyltransferase activity and suppresses hepatic glucose production independently of cell cycle progression. Through a cell-based high-throughput chemical screen, we identify a Cdk4 inhibitor that potently decreases PGC-1α acetylation. Insulin/GSK-3β (glycogen synthase kinase 3-beta) signalling induces cyclin D1 protein stability by sequestering cyclin D1 in the nucleus. In parallel, dietary amino acids increase hepatic cyclin D1 messenger RNA transcripts. Activated cyclin D1–Cdk4 kinase phosphorylates and activates GCN5, which then acetylates and inhibits PGC-1α activity on gluconeogenic genes. Loss of hepatic cyclin D1 results in increased gluconeogenesis and hyperglycaemia. In diabetic models, cyclin D1–Cdk4 is chronically elevated and refractory to fasting/feeding transitions; nevertheless further activation of this kinase normalizes glycaemia. Our findings show that insulin uses components of the cell cycle machinery in post-mitotic cells to control glucose homeostasis independently of cell division.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nakae, J., Park, B. C. & Accili, D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a wortmannin-sensitive pathway. J. Biol. Chem. 274, 15982–15985 (1999)
Cho, H. et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 292, 1728–1731 (2001)
Dentin, R. et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449, 366–369 (2007)
Matsumoto, M., Pocai, A., Rossetti, L., Depinho, R. A. & Accili, D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6, 208–216 (2007)
Puigserver, P. et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423, 550–555 (2003)
Li, X., Monks, B., Ge, Q. & Birnbaum, M. J. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature 447, 1012–1016 (2007)
Estall, J. L. et al. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-γ coactivator-1α expression. Diabetes 58, 1499–1508 (2009)
Rodgers, J. T. et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 (2005)
Lerin, C. et al. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab. 3, 429–438 (2006)
McKeehan, W. L., Adams, P. S. & Rosser, M. P. Direct mitogenic effects of insulin, epidermal growth factor, glucocorticoid, cholera toxin, unknown pituitary factors and possibly prolactin, but not androgen, on normal rat prostate epithelial cells in serum-free, primary cell culture. Cancer Res. 44, 1998–2010 (1984)
Ish-Shalom, D. et al. Mitogenic properties of insulin and insulin analogues mediated by the insulin receptor. Diabetologia 40 (suppl 2). S25–S31 (1997)
Soni, R. et al. Inhibition of cyclin-dependent kinase 4 (Cdk4) by fascaplysin, a marine natural product. Biochem. Biophys. Res. Commun. 275, 877–884 (2000)
Matsushime, H. et al. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71, 323–334 (1992)
Kato, J., Matsushime, H., Hiebert, S. W., Ewen, M. E. & Sherr, C. J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 7, 331–342 (1993)
Fry, D. W. et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 3, 1427–1438 (2004)
Boyer, L. A. et al. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10, 935–942 (2002)
Rodgers, J. T. & Puigserver, P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl Acad. Sci. USA 104, 12861–12866 (2007)
Diehl, J. A., Zindy, F. & Sherr, C. J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11, 957–972 (1997)
Hinds, P. W., Dowdy, S. F., Eaton, E. N., Arnold, A. & Weinberg, R. A. Function of a human cyclin gene as an oncogene. Proc. Natl Acad. Sci. USA 91, 709–713 (1994)
Anders, L. et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 20, 620–634 (2011)
Abella, A. et al. Cdk4 promotes adipogenesis through PPARgamma activation. Cell Metab. 2, 239–249 (2005)
Diehl, J. A., Cheng, M., Roussel, M. F. & Sherr, C. J. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511 (1998)
Choi, Y. J. et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell 22, 438–451 (2012)
Pinkert, C. A., Ornitz, D. M., Brinster, R. L. & Palmiter, R. D. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1, 268–276 (1987)
Macotela, Y. et al. Dietary leucine–an environmental modifier of insulin resistance acting on multiple levels of metabolism. PLoS ONE 6, e21187 (2011)
Wang, Y. et al. Inositol-1,4,5-trisphosphate receptor regulates hepatic gluconeogenesis in fasting and diabetes. Nature 485, 128–132 (2012)
Zeggini, E. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–1341 (2007)
Rane, S. G. et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in β-islet cell hyperplasia. Nature Genet. 22, 44–52 (1999)
Blanchet, E. et al. E2F transcription factor-1 regulates oxidative metabolism. Nature Cell Biol. 13, 1146–1152 (2011)
Schwartz, G. K. et al. Phase I study of PD 0332991, a cyclin-dependent kinase inhibitor, administered in 3-week cycles (Schedule 2/1). Br. J. Cancer 104, 1862–1868 (2011)
Li, D. et al. A modified method using TRIzol reagent and liquid nitrogen produces high-quality RNA from rat pancreas. Appl. Biochem. Biotechnol. 158, 253–261 (2009)
Vazquez, F. et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 23, 287–301 (2013)
Ogryzko, V. V. et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94, 35–44 (1998)
Rodgers, J. T., Haas, W., Gygi, S. P. & Puigserver, P. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 11, 23–34 (2010)
Jurczak, M. J. et al. Dissociation of inositol-requiring enzyme (IRE1α)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J. Biol. Chem. 287, 2558–2567(2012
Acknowledgements
We thank all the members of the Puigserver laboratory for discussions and suggestions about this project. We also appreciate the consultations with and efforts by M. Jedrychowski and S. Gygi for proteomic analysis. Y.L. was supported in part by a 21st Century Leaders scholarship from Ewha Womans University. J.E.D. was supported in part by a National Research Service Award Kirschstein Fellowship from the National Institutes of Health (NIH). The participating researchers were supported with funds from the Dana-Farber Cancer Institute and with grants from the American Diabetes Association, Department of Defense, NIH/National Institute of Diabetes and Digestive and Kidney Diseases (RO1069966 and R24DK080261-06), NIH (RO3 MH092174) awarded to P.P., NIH (RO1 CA108420) awarded to P.S. and NIH (DK059635) awarded to Yale’s Mouse Metabolic Phenotyping Center/G.I.S.
Author information
Authors and Affiliations
Contributions
P.P. and Y.L. conceived the project. Y.L. designed and performed most of the experiments aided by discussions with P.P. J.E.D. also contributed to the design and discussions of the manuscript. Additional advice in the design and execution of the experiments was provided by F.V. and J.-H.L. H.C. assisted with mice experiments. Y.J.C. and P.S. provided D1 fl/fl mice and helped with BrdU incorporation and Ki-67 staining experiments. M.J., J.P.C., H.-B.R., X.Y. and G.I.S. performed hyperinsulinemic–euglycemic clamp experiments, and analysed, interpreted and discussed the data. N.T. assisted with performance and analysis of chemical screening. Y.L. and P.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 A cell-based high-throughput screen shows compounds regulating PGC-1α acetylation.
a, Scheme of high-throughput chemical assay. b, Compounds with significant z scores either greater than 3.0 or less than −3.0 are listed. Inhibitors indicate the compounds that increased PGC-1α acetylation whereas activators indicate those that decreased PGC-1α acetylation.
Extended Data Figure 2 Cyclin D1–Cdk4 modulates PGC-1α acetylation through GCN5.
a, Fascaplysin decreases PGC-1α acetylation in a dose-dependent manner. The dose-dependent response of PGC-1α acetylation treated with fascaplysin concentrations ranging from 31.25 nM to 8 μM was analysed by western blotting. The IC50 value was calculated using three independent measurements from the assay described in Extended Data Fig. 1a. b, Chemical structures of fascaplysin and PD 0332991. c, Cdk4 is knocked down by various Cdk4 shRNAs used in Fig. 1d. d, Fascaplysin decreases PGC-1α acetylation upon EX527 or trichostatin A treatments. Cells were treated for 8 h in the case of 1 μM fascaplysin and 4 h in the case of 1 μM EX527 or trichostatin A before collection. DMSO (-) was used as a control treatment. e, Fascpalysin has blunted effect on PCAF-mediated PGC-1α acetylation. f, Ectopically expressed Cdk4 and GCN5 interact. As a comparison, PGC-1α, Sirt1 and Sirt6 were used whereas GFP was overexpressed as a negative control. g, Phosphorylation of GCN5 by cyclin D1–Cdk4 complex is reduced by fascaplysin. DMSO or 1 μM fascaplysin were added to the kinase reaction. h, In vitro phosphorylation of GST–GCN5 recombinant proteins (1–224 amino acids, 1–386 amino acids, 1–553 amino acids, 1–837 amino acids) by cyclin D1–Cdk4 and the protein level of those fragments. i, GCN5 wild type (WT), treated with fascaplysin and GCN5 T272A/S372A (AA) mutant immunoprecipitated by anti-phospho-S*P (pS*P) antibody. j, Acetylation of PGC-1α closely follows the amount of PAF65-β bound to GCN5. Nuclear extracts of U-2OS overexpressing various amounts of GCN5 were used for western blot analysis to detect GCN5 and PAF65-β. Empty vector was transfected as a negative control. k, Interaction between GCN5 T272A/S372A (AA) and PAF65-β is reduced compared with GCN5 wild type (WT). U-2OS cells were used for western blot analysis experiments.
Extended Data Figure 3 Cyclin D1–Cdk4 regulates gluconeogenesis in primary hepatocytes and in whole animals.
a, Western blot analysis of endogenous, forskolin-induced (Fsk) or adenovirally overexpressed (O/E) PGC-1α. Nuclear extracts of primary hepatocytes were used to immunoprecipitate PGC-1α. Cells were infected with GFP or PGC-1α 48 h before collection. Forskolin (10 μM) was added for 2 h before collection. b, PD 0332991 increases forskolin-induced gluconeogenic gene expression. Primary hepatocytes were treated with 10 μM forskolin for 1.5 h after 3 h of starvation medium incubation while 1 μM PD 0332991 was added overnight (one-way ANOVA with Tukey post-test, n = 3). c, Cdk4 knockdown increases forskolin-induced gluconeogenic gene expression (one-way ANOVA with Tukey post-test, n = 3). d, Cyclin D1 wild-type, but not cyclin D1 K112E mutant, suppresses forskolin-induced gluconeogenic gene expression (one-way ANOVA with Tukey post-test, n = 3). e, Phosphorylation of GCN5, FoxO1 amino (N) terminus, FoxO1 carboxy (C) terminus, FoxO3A and PGC-1α SR domain by cyclin D1–Cdk4. f, Pgc1α knockdown blocks the increase of forskolin-induced gluconeogenic genes by fascaplysin in HepG2 cells. Pgc1α knockdown or a negative control of HepG2 cells were treated with 30 μM forskolin and 1 μM fascaplysin overnight (one-way ANOVA with Tukey post-test, n = 3). g, Gcn5 knockdown blunts the increase of gluconeogenic gene expression caused by Cdk4 knockdown. Quantitative polymerase chain reaction with reverse transcription (qRT–PCR) analysis of Pck1 and Gcn5 and western blot of Cdk4 and Gcn5 knockdown are shown. All cells were infected with PGC-1α adenoviruses (one-way ANOVA with Tukey post-test, n = 15). h, PD 0332991 increases gluconeogenic genes when combined with GCN5 wild type (WT) overexpression, but not with GCN5 T272A/S372A (AA) mutant. GFP infected cells shown for comparison with GCN5 overexpressing cells. All cells were infected with PGC-1α adenoviruses (two-tailed unpaired t-test, n = 6). i, j, Insulin levels measured from serum and western blot analysis of Rb and AKT using nuclear (N) and cytoplasmic (C) liver extracts from mice treated with vehicle or 150 mg kg−1 PD 0332991, shown in Fig. 2l, m (i: two-tailed unpaired t-test, n = 18/GFP, n = 17/PD 0332991). k, Levels of cyclin D1 and Rb phosphorylation in GFP or cyclin D1 T286A tail-vein-injected mice, shown in Fig. 2n, o. Statistical significance is represented by asterisks corresponding to *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± s.e.m.
Extended Data Figure 4 PD 0332991 administration or cyclin D1 T286A adenoviral overexpression does not cause toxicity compared with its respective control treatment.
a, Basal physiological indexes of mice challenged with either vehicle or PD 0332991 administration (n = 5). b, Basal physiological indexes of mice injected with either GFP or cyclin D1 T286A adenoviruses (n = 5). (ALT, alanine transaminase; AST, aspartate transaminase; LDH, lactate dehydrogenase; mean ± s.e.m.).
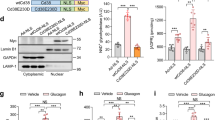
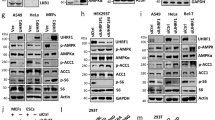
Extended Data Figure 5 Cyclin D1–Cdk4 is regulated by insulin/GSK-3β, and hepatic specific cyclin D1 deletion causes increased gluconeogenesis and glycaemia upon re-feeding.
a, Cyclin D1 transcripts are increased upon re-feeding. qRT–PCR analysis of Ccnd1, Ccnd2 and Ccne1 gene expression upon overnight fasting, 4 h and 10 h re-feeding in BALB/c mice livers (one-way ANOVA with Tukey post-test, n = 3). b, Cyclin D1 protein is increased upon re-feeding. Western blot analysis of cyclin D1 protein levels and associated signalling pathway upon fasting and re-feeding measured from nuclear (N) and cytoplasmic (C) liver extracts from BALB/c mice. c, Cyclin D1–Cdk4 kinase activity is increased upon 4 h re-feding. In vitro 32P incorporation into recombinant Rb by immunoprecipitated cyclin D1–Cdk4 kinase from whole-cell extracts of overnight fast and 4 h re-fed livers. d, Western blot analysis of cyclin D1 protein level and associated signalling pathway upon fasting and re-feeding and qRT–PCR analysis of Ccnd1 and Pgc1α mRNA level in various tissues (L, liver; M, skeletal muscle; B, brown adipose tissue; W, epididymal white adipose tissue; P, pancreas; two-tailed unpaired t-test, n = 12/L, n = 4/M, B, W, P). e, qRT–PCR analysis of PGC-1α target genes in liver and epididymal white adipose tissues (eWAT) upon vehicle or PD 0332991 treatment (two-tailed unpaired t-test, n = 10). f, Ccnb1 and Pcna gene expressions in liver do not change upon fasting and re-feeding (n = 3/fast and 10 h re-fed, n = 4/4 h re-fed). g, BrdU incorporation in liver does not change upon fasting and re-feeding. Small intestine was used as a positive control. h, Ki-67 staining in liver does not change upon fasting and re-feeding after vehicle or PD 0332991 administration. The small intestine was used as a positive control. i, Ki-67 staining in liver does not change upon fasting and re-feeding following GFP or cyclin D1 T286A tail-vein injection. Small intestine used as a positive control. j, Hepatic ploidy profiles of livers of GFP or cyclin D1 T286A adenovirus tail-vein injected mice do not show significant difference. Ploidy analysis of primary hepatocytes isolated from livers measured by propidium iodide staining and flow cytometry (n = 6/fast and 4 h re-fed, n = 4/10 h re-fed). k, Western blot analysis of endogenous nuclear (N) and cytoplasmic (C) cyclin D1 protein level upon insulin or GSK-3β inhibitors treatments in primary hepatocytes. l, PGC-1α acetylation is increased upon insulin or GSK-3β inhibitors treatment in primary hepatocytes. m, No effect of insulin or GSK-3β inhibitors on cyclin D1 mRNA level (n = 3). n, Minimum essential medium (MEM) amino acid addition increases cyclin D1 mRNA in primary hepatocytes (one-way ANOVA with Tukey post-test, n = 3). o, Insulin does not change Ccnd1 mRNA in primary hepatocytes (one-way ANOVA with Tukey post-test, n = 3). p, q, Body and liver weights and Ccnd1 and Ccnd2 gene expression of wild-type and liver-specific cyclin D1 LKO mice (combined four cohorts of n = 3/fasting, n = 5/re-feeding). r, Western blot analysis of cyclin D1 protein levels and associated signalling pathway by using nuclear and cytoplasmic liver extracts from wild-type and D1 LKO mice upon fasting (F) and 4 h re-feeding (R). s, Endogenous acetylation of PGC-1α is decreased in livers of D1 LKO mice compared with wild-type mice. Western blot analysis of acetylation of PGC-1α immunoprecipitated from liver nuclear extracts. All mice were killed upon 4 h re-feeding. t, u, PD 0332991 increases glycaemia with a similar tendency for gluconeogenic gene expression only in wild-type mice, but not in D1 LKO mice (two-tailed unpaired t-test, n = 8, except n = 6 for vehicle treated wild-type mice). v, w, Gluconeogenic gene expression and hepatic glucose production are increased in primary hepatocytes isolated from D1 LKO mice (v: one-way ANOVA Tukey post-test, n = 3; w: two-tailed unpaired test, n = 6). Statistical significance is represented by asterisks corresponding to *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± s.e.m.
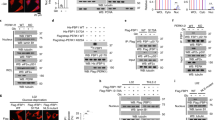
Extended Data Figure 6 In diabetic hyperinsulinemic mice, cyclin D1–Cdk4 is dysregulated and hyperactivation of cyclin D1–Cdk4 attenuates the diabetic phenotype.
a, qRT–PCR analysis of gluconeogenic and Ccnd1 gene expression changes upon fasting and re-feeding in Leprdb/+ (db/+) and Leprdb/db (db/db) mice livers (two-tailed unpaired t-test, n = 3). b, Cyclin D1 protein is chronically elevated upon fasting and re-feeding in livers of mice fed a high-fat diet compared with control diet fed mice. Nuclear (N) and cytoplasmic (C) liver extracts were used. c, d, Phosphorylation of GCN5 is elevated upon fasting in db/db of livers of mice fed a high-fat diet (HFD) compared with respective control mice and remains insensitive to fast–re-fed transitions. Western blot analysis of GCN5 immunoprecipitated by anti-phosphoS*P (pS*P) antibody using liver extracts from mice that were tail-vein injected with adenoviruses expressing GFP or GCN5 (F, 16 h fast; R, 4 h re-fed). e, Cyclin D1 and Rb phosphorylation levels in livers from db/db mice tail-vein injected with GFP, cyclin D1 T286A or cyclin D1 K112E adenoviruses, shown in Fig. 4b, c. Nuclear and cytoplasmic liver extracts were used. f, g, Cyclin D1 T286A overexpression reduces gluconeogenic genes and glycaemia in mice fed a high-fat diet (two-tailed unpaired t-test, n = 6/GFP, n = 7/D1 T286A). h, Cyclin D1 and Rb phosphorylation levels in livers of mice fed a high-fat diet that were tail-vein injected with adenoviruses expressing GFP or cyclin D1 T286A, shown in Extended Data Fig. 4f, g. i, Overall model. Statistical significance is represented by asterisks corresponding to *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as mean ± s.e.m.
Rights and permissions
About this article
Cite this article
Lee, Y., Dominy, J., Choi, Y. et al. Cyclin D1–Cdk4 controls glucose metabolism independently of cell cycle progression. Nature 510, 547–551 (2014). https://doi.org/10.1038/nature13267
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13267
This article is cited by
-
CDK4/6 inhibitor-induced bone marrow micronuclei might be caused by cell cycle arrest during erythropoiesis
Genes and Environment (2024)
-
A transient increase of HIF-1α during the G1 phase (G1-HIF) ensures cell survival under nutritional stress
Cell Death & Disease (2023)
-
Therapeutic potential of CDK4/6 inhibitors in renal cell carcinoma
Nature Reviews Urology (2022)
-
Posttranslational modifications in diabetes: Mechanisms and functions
Reviews in Endocrine and Metabolic Disorders (2022)
-
A Cdk4/6-dependent phosphorylation gradient regulates the early to late G1 phase transition
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.