Abstract
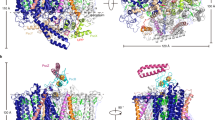
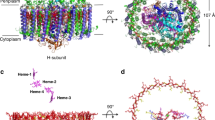
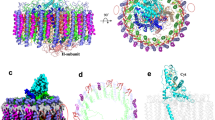
The light-harvesting core antenna (LH1) and the reaction centre (RC) of purple photosynthetic bacteria form a supramolecular complex (LH1–RC) to use sunlight energy in a highly efficient manner. Here we report the first near-atomic structure, to our knowledge, of a LH1–RC complex, namely that of a Ca2+-bound complex from Thermochromatium tepidum, which reveals detailed information on the arrangement and interactions of the protein subunits and the cofactors. The RC is surrounded by 16 heterodimers of the LH1 αβ-subunit that form a completely closed structure. The Ca2+ ions are located at the periplasmic side of LH1. Thirty-two bacteriochlorophyll and 16 spirilloxanthin molecules in the LH1 ring form an elliptical assembly. The geometries of the pigment assembly involved in the absorption characteristics of the bacteriochlorophyll in LH1 and excitation energy transfer among the pigments are reported. In addition, possible ubiquinone channels in the closed LH1 complex are proposed based on the atomic structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Deisenhofer, J., Epp, O., Miki, K., Huber, R. & Michel, H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature 318, 618–624 (1985)
Allen, J. P., Feher, G., Yeates, T. O., Komiya, H. & Rees, D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: the protein subunits. Proc. Natl Acad. Sci. USA 84, 6162–6166 (1987)
Nogi, T., Fathir, I., Kobayashi, M., Nozawa, T. & Miki, K. Crystal structures of photosynthetic reaction center and high-potential iron-sulfur protein from Thermochromatium tepidum: thermostability and electron transfer. Proc. Natl Acad. Sci. USA 97, 13561–13566 (2000)
McDermott, G. et al. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature 374, 517–521 (1995)
Koepke, J., Hu, X., Muenke, C., Schulten, K. & Michel, H. The crystal structure of the light-harvesting complex II (B800–850) from Rhodospirillum molischianum. Structure 4, 581–597 (1996)
McLuskey, K., Prince, S. M., Cogdell, R. J. & Isaacs, N. W. The crystallographic structure of the B800-820 LH3 light-harvesting complex from the purple bacteria Rhodopseudomonas acidophila strain 7050. Biochemistry 40, 8783–8789 (2001)
Karrasch, S., Bullough, P. A. & Ghosh, R. The 8.5 Å projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J. 14, 631–638 (1995)
Jamieson, S. J. et al. Projection structure of the photosynthetic reaction centre-antenna complex of Rhodospirillum rubrum at 8.5 Å resolution. EMBO J. 21, 3927–3935 (2002)
Scheuring, S. et al. Nanodissection and high-resolution imaging of the Rhodopseudomonas viridis photosynthetic core complex in native membranes by AFM. Proc. Natl Acad. Sci. USA 100, 1690–1693 (2003)
Scheuring, S. et al. Structural role of PufX in the dimerization of the photosynthetic core complex of Rhodobacter sphaeroides. J. Biol. Chem. 279, 3620–3626 (2004)
Jungas, C., Ranck, J. L., Rigaud, J. L., Joliot, P. & Verméglio, A. Supramolecular organization of the photosynthetic apparatus of Rhodobacter sphaeroides. EMBO J. 18, 534–542 (1999)
Siebert, C. A. et al. Molecular architecture of photosynthetic membranes in Rhodobacter sphaeroides: the role of PufX. EMBO J. 23, 690–700 (2004)
Qian, P., Hunter, C. N. & Bullough, P. A. The 8.5 Å projection structure of the core RC-LH1-PufX dimer of Rhodobacter sphaeroides. J. Mol. Biol. 349, 948–960 (2005)
Scheuring, S., Busselez, J. & Lévy, D. Structure of the dimeric PufX-containing core complex of Rhodobacter blasticus by in situ atomic force microscopy. J. Biol. Chem. 280, 1426–1431 (2005)
Roszak, A. W. et al. Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science 302, 1969–1972 (2003)
Richter, M. F. et al. Refinement of the X-ray structure of the RC-LH1 core complex from Rhodopseudomonas palustris by single-molecule spectroscopy. Proc. Natl Acad. Sci. USA 104, 20280–20284 (2007)
Suzuki, H. et al. Purification, characterization and crystallization of the core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum. Biochim. Biophys. Acta 1767, 1057–1063 (2007)
Madigan, M. T. A novel photosynthetic purple bacterium isolated from a Yellowstone hot spring. Science 225, 313–315 (1984)
Rowe, J. J., Fournier, R. O. & Morey, G. W. Chemical analysis of thermal waters in Yellowstone National Park, Wyoming, 1960–65. U.S. Geological Survey Bulletin 1303 http://pubs.usgs.gov/bul/1303/report.pdf (1973)
Kimura, Y. et al. Calcium ions are involved in the unusual red shift of the light-harvesting 1 Qy transition of the core complex in thermophilic purple sulfur bacterium Thermochromatium tepidum. J. Biol. Chem. 283, 13867–13873 (2008)
Kimura, Y., Yu, L. J., Hirano, Y., Suzuki, H. & Wang, Z.-Y. Calcium ions are required for the enhanced thermal stability of the light-harvesting-reaction center core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum. J. Biol. Chem. 284, 93–99 (2009)
Yu, L. J., Kato, S. & Wang, Z.-Y. Examination of the putative Ca2+-binding site in the light-harvesting complex 1 of thermophilic purple sulfur bacterium Thermochromatium tepidum. Photosynth. Res. 106, 215–220 (2010)
Papiz, M. Z., Prince, S. M., Howard, T., Cogdell, R. J. & Isaacs, N. W. The structure and thermal motion of the B800–850 LH2 complex from Rps. acidophila at 2.0 Å resolution and 100 K: new structural features and functionally relevant motions. J. Mol. Biol. 326, 1523–1538 (2003)
Fathir, I. et al. Biochemical and spectral characterization of the core light harvesting complex 1 (LH1) from the thermophilic purple sulfur bacterium Chromatium tepidum. Photosynth. Res. 58, 193–202 (1998)
Kimura, Y. et al. Metal cations modulate the bacteriochlorophyll–protein interaction in the light-harvesting 1 core complex from Thermochromatium tepidum. Biochim. Biophys. Acta 1817, 1022–1029 (2012)
Davis, C. M., Bustamante, P. L. & Loach, P. A. Reconstitution of the bacterial core light-harvesting complexes of Rhodobacter sphaeroides and Rhodospirillum rubrum with isolated α- and β-polypeptides, bacteriochlorophyll a, and carotenoid. J. Biol. Chem. 270, 5793–5804 (1995)
Walz, T. & Ghosh, R. Two-dimensional crystallization of the light-harvesting I-reaction centre photounit from Rhodospirillum rubrum. J. Mol. Biol. 265, 107–111 (1997)
Bahatyrova, S. et al. Flexibility and size heterogeneity of the LH1 light harvesting complex revealed by atomic force microscopy. J. Biol. Chem. 279, 21327–21333 (2004)
Scheuring, S. in The purple phototrophic bacteria (eds Hunter, C. N., Daldal, F. & Beatty, J. T. ) 941–952 (Springer, 2009)
Comayras, F., Jungas, C. & Lavergne, J. Functional consequences of the organization of the photosynthetic apparatus in Rhodobacter sphaeroides: II. A study of PufX- membranes. J. Biol. Chem. 280, 11214–11223 (2005)
Mascle-Allemand, C., Lavergne, J., Bernadac, A. & Sturgis, J. N. Organisation and function of the Phaeospirillum molischianum photosynthetic apparatus. Biochim. Biophys. Acta 1777, 1552–1559 (2008)
Aird, A., Wrachtrup, J., Schulten, K. & Tietz, C. Possible pathway for ubiquinone shuttling in Rhodospirillum rubrum revealed by molecular dynamics simulation. Biophys. J. 92, 23–33 (2007)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997)
Collaborative Computational Project, Number 4. The CCP4suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Terwilliger, T. SOLVE and RESOLVE: automated structure solution, density modification, and model building. J. Synchrotron Radiat. 11, 49–52 (2004)
Cowtan, K. ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Prot. Crystallogr. 31, 34–38 (1994)
McRee, D. E. XtalView/Xfit - a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Haddadian, E. J. et al. Automated real-space refinement of protein structure using a realistic backbone move set. Biophys. J. 101, 899–909 (2011)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK - a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993)
DeLano, W. L. The PyMOL Molecular Graphics System DeLano Scientific, San Carlos, CA,. http://www.pymol.org. (2002)
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers 22, 2577–2637 (1983)
Fathir, I. et al. The genes coding for the L, M and cytochrome subunits of the photosynthetic reaction center from the thermophilic purple sulfur bacterium Chromatium tepidum. Photosynth. Res. 51, 71–82 (1997)
Acknowledgements
We thank S. Takaichi for analysing the pigment compositions in LH1–RC and dry cells; F. Sekine for sequencing the genes encoding RC proteins; T. Nozawa for his support and interest at the initial stage; Y. Kimura, Masayuki Kobayashi and J.-R. Shen for their discussion; H. Suzuki, Dorina, M. Nakamura, F. Oh-hata and Miho Kobayashi for their contributions to the early stage of crystallization and data collection, and the Kao Corporation for kindly providing LDAO. This work was supported by a Grant-in-Aid for Scientific Research on the Priority Area “Structures of Biological Macromolecular Assemblies” and a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; by funds from the Takeda Science Foundation, the Kurata Memorial Hitachi Science and Technology Foundation (to Z.-Y.W.-O.), and the Targeted Proteins Research Program and the Photon and Quantum Basic Research Coordinated Development Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.M.), and by a postdoctoral fellowship from Ibaraki University (to L.-J.Y.). This work was performed using the synchrotron beamline BL44XU at SPring-8 under the Cooperative Research Program of the Institute for Protein Research, Osaka University and the beamlines at the KEK Photon Factory. We are grateful to A. Nakagawa, E. Yamashita, N. Matsugaki and Y. Yamada for their assistance in the data collection.
Author information
Authors and Affiliations
Contributions
Z.-Y.W.-O. and K.M. initiated the project and supervised all experiments. L.-J.Y. carried out experiments of purification, crystallization and post-crystallization treatment. Y.H. conducted screenings of crystallization condition. L.-J.Y., Y.H. and T.K. collected X-ray diffraction data. S.N. and K.T. carried out structure determination. All authors contributed to the data analysis. S.N., K.T., L.-J.Y., Z.-Y.W.-O. and K.M. prepared the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Crystal packing of the LH1–RC complex.
a, The LH1–RC complexes in the crystal lattice of the P21 crystals are shown as ribbons. Molecules in the asymmetric unit are coloured in blue and light blue, whereas symmetry-related molecules are in grey. The unit cell boundaries are defined by red lines. b, The LH1–RC complexes in the crystal lattice of the C2 crystals are shown as ribbons. A molecule in the asymmetric unit is coloured in blue, whereas symmetry-related molecules are in grey.
Extended Data Figure 2 Close-up views of the electron density maps of the LH1–RC complex.
a–d, The experimental (left) and sigmaA-weighted 2Fobs − Fcalc (right) electron density maps for the transmembrane regions of RC (a) and LH1 (b), the special-pair BChls (c) and the ubiquinone molecule (d) are represented as grey meshes. The contour levels are 1.0σ except the experimental maps of c and d contoured at 0.8σ.
Extended Data Figure 3 Primary and three-dimensional structure of the RC from T. tepidum.
a, Full amino acid sequences of the T. tepidum RC proteins deduced from the re-verified nucleotide sequences of puf operon and puhA. Red shaded letters denote the corrected amino acid residues that are different from those reported previously. The letters coloured in grey denote the amino acid residues that are not visible in the electron density map. b, Comparison of the RC structures from T. tepidum. The RC in the isolated state (grey) and the RC in the LH1–RC complex (red) are superimposed. c, The most deviated portion (residues 172–196 in Cyt subunit).
Extended Data Figure 4 Anomalous diffraction signals.
a, Anomalous difference Fourier maps calculated from the data sets collected at 2.70 Å (higher energy than the K-edge of calcium) are represented as meshes in red (5σ) and pink (3σ) for the LH1–RC structure. b, A close-up view for the calcium ion located on the interface of the Cyt and M subunits at contour levels of 5σ. c, A close-up view for a calcium ion located in the LH1 at contour levels of 5σ. d, Anomalous difference Fourier maps at 3.15 Å (lower energy than the K-edge of calcium) are represented in pink at contour levels of 3σ.
Extended Data Figure 5 Sequences of α- and β-apoproteins.
a, Sequence alignment for the α-apoproteins. Fully, highly and relatively conserved residues are shaded by red, light red and pale red, respectively. The helices for the α-apoprotein of T. tepidum are represented as blue cylinders and the loops are represented as yellow lines. Unmodelled regions are represented as grey dotted lines. The purple and pink triangles indicate the coordinating residues to magnesium and calcium in the LH1 of T. tepidum, respectively. b, Sequence alignment for the β-apoproteins. The helix for the β-apoprotein of T. tepidum is represented as a green cylinder. c, Schematic representation of the α-apoprotein from T. tepidum. The filled circles in purple and pink indicate the residues coordinating to magnesium and calcium, respectively. d, Schematic representation of the β-apoprotein.
Extended Data Figure 6 Fluctuation of the extramembrane regions of the LH1.
a, The superimposition of all 16 αβ-heterodimers is shown in a stereo view. The α- and β-apoproteins are represented as blue and green tubes, respectively. The B915s are represented as purple sticks. b, The experimental (left) and sigmaA-weighted 2Fobs − Fcalc (right) electron density maps for the extramembrane regions of LH1. Main chains of the regions can be traced, whereas some side chains are difficult to observe. c, Thermal motion for Cα atoms of the LH1–RC complex are shown as thermal ellipsoids which are coloured according to the equivalent isotropic B-factors ranging from blue (smallest) to red (largest).
Extended Data Figure 7 Comparison with other antenna complexes.
a, An αβ-heterodimer of the T. tepidum LH1 (magenta) is compared with αβ-heterodimers of Rhodops. acidophila LH2 (blue). b, A view rotated by 90° from a around the vertical axis. c, An αβ-heterodimer of the LH1 of T. tepidum (magenta) is compared with αβ-heterodimers of LH2 from Rhodosp. molischianum (dark blue). d, A view rotated by 90° from c around the vertical axis. e, An αβ-heterodimer of the T. tepidum LH1 (magenta) is compared with αβ-heterodimers of Rhodops. acidophila LH3 (cyan). f, A view rotated by 90° from e around the vertical axis. g, Comparison of the ring structures of LH1 of T. tepidum (magenta) with the LH2 from Rhodosp. molischianum (dark blue), LH2 from Rhodops. acidophila (blue), and LH3 of Rhodops. acidophila (cyan). h, Comparison of the ring structure with Rhodops. palustris LH1–RC. The T. tepidum LH1–RC is coloured, whereas the complex from Rhodops. palustris is shown in grey and black (helix W).
Extended Data Figure 8 Variations in the inter-subunit spacing of LH1.
a, A cross section of the ring at the periplasmic (left panel) and cytoplasmic (right panel) ends of the transmembrane helices. Distances between the two neighbouring helices are indicated by the black lines connecting them. For the α-apoproteins, the distances are measured between the Cα atoms of the two neighbouring α-Val 39 at the periplasmic end and the two neighbouring α-Pro 17 at the cytoplasmic end. In a similar way, the distances are measured between the Cα atoms of the two neighbouring β-Ala 39 at the periplasmic end and the two neighbouring β-Phe 17 at the cytoplasmic end for the β-apoproteins. b, The putative quinone channels are viewed on a cross section of the molecular surface from the periplasmic side. The head groups of the ubiquinone molecule at the QB site are represented as red CPK models. c, Cross sections of two channels in the LH1 with different shapes are superimposed. The molecular envelopes of the widest and narrowest channels are represented in blue and pale red, respectively. The red arrow illustrates the diffusion route for the ubiquinones.
Rights and permissions
About this article
Cite this article
Niwa, S., Yu, LJ., Takeda, K. et al. Structure of the LH1–RC complex from Thermochromatium tepidum at 3.0 Å. Nature 508, 228–232 (2014). https://doi.org/10.1038/nature13197
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13197
This article is cited by
-
Atomic force microscopic analysis of the light-harvesting complex 2 from purple photosynthetic bacterium Thermochromatium tepidum
Photosynthesis Research (2023)
-
A self-complementary macrocycle by a dual interaction system
Nature Communications (2022)
-
Complete genome of the thermophilic purple sulfur Bacterium Thermochromatium tepidum compared to Allochromatium vinosum and other Chromatiaceae
Photosynthesis Research (2022)
-
Exciton transfer between LH1 antenna complex and photosynthetic reaction center dimer
Journal of Biological Physics (2021)
-
Cryo-EM structure of a Ca2+-bound photosynthetic LH1-RC complex containing multiple αβ-polypeptides
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.