Abstract
Successful mammalian cloning using somatic cell nuclear transfer (SCNT) into unfertilized, metaphase II (MII)-arrested oocytes attests to the cytoplasmic presence of reprogramming factors capable of inducing totipotency in somatic cell nuclei1,2,3. However, these poorly defined maternal factors presumably decline sharply after fertilization, as the cytoplasm of pronuclear-stage zygotes is reportedly inactive4,5. Recent evidence suggests that zygotic cytoplasm, if maintained at metaphase, can also support derivation of embryonic stem (ES) cells after SCNT6,7,8, albeit at low efficiency. This led to the conclusion that critical oocyte reprogramming factors present in the metaphase but not in the interphase cytoplasm are ‘trapped’ inside the nucleus during interphase and effectively removed during enucleation9. Here we investigated the presence of reprogramming activity in the cytoplasm of interphase two-cell mouse embryos (I2C). First, the presence of candidate reprogramming factors was documented in both intact and enucleated metaphase and interphase zygotes and two-cell embryos. Consequently, enucleation did not provide a likely explanation for the inability of interphase cytoplasm to induce reprogramming. Second, when we carefully synchronized the cell cycle stage between the transplanted nucleus (ES cell, fetal fibroblast or terminally differentiated cumulus cell) and the recipient I2C cytoplasm, the reconstructed SCNT embryos developed into blastocysts and ES cells capable of contributing to traditional germline and tetraploid chimaeras. Last, direct transfer of cloned embryos, reconstructed with ES cell nuclei, into recipients resulted in live offspring. Thus, the cytoplasm of I2C supports efficient reprogramming, with cell cycle synchronization between the donor nucleus and recipient cytoplasm as the most critical parameter determining success. The ability to use interphase cytoplasm in SCNT could aid efforts to generate autologous human ES cells for regenerative applications, as donated or discarded embryos are more accessible than unfertilized MII oocytes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A. J. & Campbell, K. H. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 (1997)
Gurdon, J. B. & Wilmut, I. Nuclear transfer to eggs and oocytes. Cold Spring Harb. Perspect. Biol. 3, a002659 (2011)
Tachibana, M. et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–1238 (2013)
McGrath, J. & Solter, D. Inability of mouse blastomere nuclei transferred to enucleated zygotes to support development in vitro . Science 226, 1317–1319 (1984)
Wakayama, T., Tateno, H., Mombaerts, P. & Yanagimachi, R. Nuclear transfer into mouse zygotes. Nature Genet. 24, 108–109 (2000)
Egli, D., Sandler, V. M., Shinohara, M. L., Cantor, H. & Eggan, K. Reprogramming after chromosome transfer into mouse blastomeres. Curr. Biol. 19, 1403–1409 (2009)
Egli, D., Rosains, J., Birkhoff, G. & Eggan, K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature 447, 679–685 (2007)
Riaz, A. et al. Mouse cloning and somatic cell reprogramming using electrofused blastomeres. Cell Res. 21, 770–778 (2011)
Egli, D., Birkhoff, G. & Eggan, K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nature Rev. Mol. Cell Biol. 9, 505–516 (2008)
Nichols, J. et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998)
Zhang, J. et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nature Cell Biol. 8, 1114–1123 (2006)
Festuccia, N. et al. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 11, 477–490 (2012)
Jullien, J., Pasque, V., Halley-Stott, R. P., Miyamoto, K. & Gurdon, J. B. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nature Rev. Mol. Cell Biol. 12, 453–459 (2011)
Zheng, L., Roeder, R. G. & Luo, Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 (2003)
Wakayama, T., Rodriguez, I., Perry, A. C., Yanagimachi, R. & Mombaerts, P. Mice cloned from embryonic stem cells. Proc. Natl Acad. Sci. USA 96, 14984–14989 (1999)
Mitalipov, S. M. et al. Reprogramming following somatic cell nuclear transfer in primates is dependent upon nuclear remodeling. Hum. Reprod. 22, 2232–2242 (2007)
Jaroudi, S. & SenGupta, S. DNA repair in mammalian embryos. Mutat. Res. 635, 53–77 (2007)
Schöler, H. R., Dressler, G. R., Balling, R., Rohdewohld, H. & Gruss, P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 9, 2185–2195 (1990)
Zambrowicz, B. P. et al. Disruption of overlapping transcripts in the ROSA βgeo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA 94, 3789–3794 (1997)
Wu, G. et al. Generation of healthy mice from gene-corrected disease-specific induced pluripotent stem cells. PLoS Biol. 9, e1001099 (2011)
Kishigami, S. et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 340, 183–189 (2006)
Tsunoda, Y. et al. Full-term development of mouse blastomere nuclei transplanted into enucleated two-cell embryos. J. Exp. Zool. 242, 147–151 (1987)
Eckardt, S., Leu, N. A., Kurosaka, S. & McLaughlin, K. J. Differential reprogramming of somatic cell nuclei after transfer into mouse cleavage stage blastomeres. Reproduction 129, 547–556 (2005)
Mitalipov, S. M., Yeoman, R. R., Nusser, K. D. & Wolf, D. P. Rhesus monkey embryos produced by nuclear transfer from embryonic blastomeres or somatic cells. Biol. Reprod. 66, 1367–1373 (2002)
Stice, S. L. & Robl, J. M. Nuclear reprogramming in nuclear transplant rabbit embryos. Biol. Reprod. 39, 657–664 (1988)
Willadsen, S. M. Nuclear transplantation in sheep embryos. Nature 320, 63–65 (1986)
Cheong, H. T., Takahashi, Y. & Kanagawa, H. Birth of mice after transplantation of early cell-cycle-stage embryonic nuclei into enucleated oocytes. Biol. Reprod. 48, 958–963 (1993)
Stice, S. L., Keefer, C. L. & Matthews, L. Bovine nuclear transfer embryos: oocyte activation prior to blastomere fusion. Mol. Reprod. Dev. 38, 61–68 (1994)
Egli, D. et al. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nature Commun. 2, 488 (2011)
Lee, H. S. et al. Rapid mitochondrial DNA segregation in primate preimplantation embryos precedes somatic and germline bottleneck. Cell. Rep. 1, 506–515 (2012)
Acknowledgements
The authors would like to acknowledge the Small Lab Animal Unit at Oregon National Primate Research Center for providing expertise and services that contributed to this project. We are indebted to E. Wolff, A. Sugawara, C. Ramsey, H.-S. Lee, J. Woodward, D. Melquizo Sanchis, C. Van Dyken and B. Manson for their technical support. The study was supported by grants from the National Institutes of Health (R01HD063276, R01HD057121, R01HD059946, R01EY021214, P51OD011092) and funds from the Leducq Fondation and the Collins Medical Trust.
Author information
Authors and Affiliations
Contributions
S.M. and E.K. conceived and planned the study, E.K. carried out SCNT experiments, G.W. conducted tetraploid chimera experiments, H.M., Y.L., R.T.-H., M.T. and M.S. assisted with colony management, oocyte and embryo collections, embryo transfers, ES cell isolation and culture and expression profiling. E.K., D.P.W., H.R.S. and S.M. analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
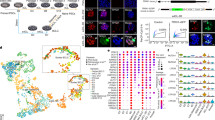
Extended Data Figure 1 Maternal and embryonic gene expression patterns.
a, Immunocytochemical detection of Gapdh signal demonstrated even distribution in nuclei and cytoplasm in MII oocytes and interphase zygotes and two-cell embryos (original magnification, ×400). b, Expression of Gapdh normalized to Actb. No significant differences were seen between intact and enucleated two-cell embryos (four replicates each containing pooled RNA from five embryos, P > 0.05). c, Gapdh expression was relatively low and constant with no significant differences seen until the eight-cell stage. Expression was increased in 16-cell embryos and blastocysts (three replicates each containing pooled RNA from ten embryos, *P < 0.05; **P < 0.01). d, Gene expression of the transcriptional regulators Bmi1, Hsf1, Brg1, Sall4 and Esrrb, the transcription factor Oct4, and epigenetic factors Apobec1, Aid and Tet1 during mouse pre-implantation embryo development. The level of expression of Brg1, Apobec1, Oct4, Sall4, Aid, Tet1 and Esrrb underwent marked increases at or after the four-cell stage. The red bars indicate enucleated oocytes, zygotes or two-cell embryos. No significant differences were observed between intact and enucleated counterparts (n = 3 biological replicates, P > 0.05). Gene expression was normalized to Gapdh. Error bars indicate average ± s.d. cyto, cytoplast; I, interphase; M, metaphase; MII, metaphase II-arrested oocyte; Z, zygote.
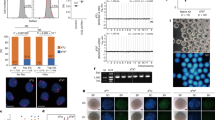
Extended Data Figure 2 Protein expression of candidate reprogramming factors.
a, Immunocytochemical detection of Bmi1, Hsf1 and Brg1 in zygotic interphase, metaphase and two-cell interphase embryos. The expression pattern of these proteins was similar to that observed for mRNA expression (Extended Data Fig. 1d). I2C embryos were fixed 4 h after cleavage. I, interphase; M, metaphase; Z, zygote. b, Western blot detection of Brg1 in nuclear donor cells and in intact and enucleated two-cell embryos (n = 40). c, Immunocytochemical detection of Brg1 and Hsf1 in nuclear donor FFs and ES cells. Scale bars, 400 µm. d, Immunocytochemical detection of Brg1 and Hsf1 in SCNT embryos (original magnification, ×400). MII, MII oocyte.
Extended Data Figure 3 Cell cycle determination in nuclear donor cells.
a, Confluent FF populations subjected to flow cytometric processing. After sorting by size and density, 91% of small-sized cells were in the G0/G1 phase of the cell cycle. b, Proliferating FF populations were mostly at S phase. c, ES cell populations by flow cytometry. The size was correlated with the cell cycle, 70% of the small-, medium- and large-sized cells were at G0/G1, S and G2/M, respectively. d, FITC-conjugated BrdU was used to define S-phase FFs and ES cells. PI, propidium iodide. Scale bars, 100 μm. e, Correlation of cell size and cell cycle in FFs and ES cells. Cell size was measured photographically after 3.7% formaldehyde fixation. S-phase cells were separated by BrdU integration (green). The assignments of cell size provided in panels a, b and c were defined from the results in panel e (*P < 0.05). pFFs, proliferating fetal fibroblasts.
Extended Data Figure 4 Nuclear staining of cell-cycle-matched and mismatched SCNT embryos.
Left, expanded nuclei of developmentally competent two-cell SCNT embryos generated by transfer of S-phase FF nuclei into enucleated S-phase two-cell embryo. Middle and right panels, condensed or dispersed nuclei of arrested SCNT embryos generated after cell cycle mismatch between donor nucleus and recipient cytoplasm. Embryos were arrested and apoptotic by the G1/S or S/G2 cell cycle checkpoints, respectively. SCNT embryos were fixed 15 h after cleavage (original magnification, ×400).
Extended Data Figure 5 Genotyping of chimaeric mice.
a, Chromatogram of mtDNA depicting sequence differences at 11,939, 12,062 and 12,122 base pairs (bp) and demonstrating that mtDNA in ntES cells was derived from recipient embryo cytoplasm. Sequence differences between chimaera host embryos and ntES cells at 11,939, 12,199, 12,002, 12,005, 12,008 and 12,062 bp allowed quantification of chimaerism. The mtDNA from a diploid chimaeric mouse appeared as a double peak. The tetraploid chimaera showed only ntES cell mtDNA (GeneBank accession EF108343.1). b, Allele refractory mutation system quantitative PCR (ARMS-qPCR) assay in diploid and tetraploid chimaeras. Similar colours indicate similar origin. Distances between colour plots represent the contribution of chimaeric host embryos. Amplification plot (left) and bar graph (right). Detection limit of this assay is 0.01%. c, High ntES cell contribution chimaeric female and her pups demonstrating germline transmission. White spot on the head of the dam originated from the ICR host embryo. d, Tetraploid embryo complementation pups born after ntES cell (black) injection into host eight-cell-stage tetraploid embryos. e, Genotyping of tetraploid complementation pups.
Extended Data Figure 6 Cloned offspring and genotyping.
a, Body and placenta of cloned and control pups delivered by caesarean section at 20 days. Clones and placentas were larger or smaller than controls. Scale bars, 0.5 cm. b, Fetal and placental weights (g) of cloned and control pups. c, Genotyping of nuclear donor cells and cloned pups carrying ROSA26+/−OG2+/−.
Supplementary information
Enucleation and nuclear transfer procedures developed for interphase SCNT into 2-cell mouse embryos
A mouse 2-cell embryo was secured with holding pipette and zona pellucida was drilled with a laser pulse and then an enucleation pipette was inserted through the opening. Both blastomeres were enucleated under Hoffman contrast microscopy, and an intact nuclear donor cell was introduced into each enucleated blastomere using fusogenic activity of HVJ-E. (WMV 12050 kb)
Rights and permissions
About this article
Cite this article
Kang, E., Wu, G., Ma, H. et al. Nuclear reprogramming by interphase cytoplasm of two-cell mouse embryos. Nature 509, 101–104 (2014). https://doi.org/10.1038/nature13134
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13134
This article is cited by
-
Haploidy in somatic cells is induced by mature oocytes in mice
Communications Biology (2022)
-
Wandering along the epigenetic timeline
Clinical Epigenetics (2020)
-
Long non-coding RNAs potentially function synergistically in the cellular reprogramming of SCNT embryos
BMC Genomics (2018)
-
Genomic instability during reprogramming by nuclear transfer is DNA replication dependent
Nature Cell Biology (2017)
-
Cytoplasmic Determination of Meiotic Spindle Size Revealed by a Unique Inter-Species Germinal Vesicle Transfer Model
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.