Abstract
The detection of microbial pathogens involves the recognition of conserved microbial components by host cell sensors such as Toll-like receptors (TLRs) and NOD-like receptors (NLRs). TLRs are membrane receptors that survey the extracellular environment for microbial infections, whereas NLRs are cytosolic complexes that detect microbial products that reach the cytosol. Upon detection, both sensor classes trigger innate inflammatory responses and allow the engagement of adaptive immunity1,2. Endo-lysosomes are the entry sites for a variety of pathogens, and therefore the sites at which the immune system first senses their presence. Pathogens internalized by endocytosis are well known to activate TLRs 3 and 7–9 that are localized to endocytic compartments and detect ligands present in the endosomal lumen3. Internalized pathogens also activate sensors in the cytosol such as NOD1 and NOD2 (ref. 2), indicating that endosomes also provide for the translocation of bacterial components across the endosomal membrane. Despite the fact that NOD2 is well understood to have a key role in regulating innate immune responses and that mutations at the NOD2 locus are a common risk factor in inflammatory bowel disease and possibly other chronic inflammatory states4,5, little is known about how its ligands escape from endosomes. Here we show that two endo-lysosomal peptide transporters, SLC15A3 and SLC15A4, are preferentially expressed by dendritic cells, especially after TLR stimulation. The transporters mediate the egress of bacterially derived components, such as the NOD2 cognate ligand muramyl dipeptide (MDP)6,7, and are selectively required for NOD2 responses to endosomally derived MDP. Enhanced expression of the transporters also generates endosomal membrane tubules characteristic of dendritic cells, which further enhanced the NOD2-dependent response to MDP. Finally, sensing required the recruitment of NOD2 and its effector kinase RIPK2 (refs 8, 9) to the endosomal membrane, possibly by forming a complex with SLC15A3 or SLC15A4. Thus, dendritic cell endosomes are specialized platforms for both the lumenal and cytosolic sensing of pathogens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
01 January 2014
: In Fig. 1f, ‘Mouse Slc15a4’ was incorrectly given as ‘Mouse Slc15a3’, and has been fixed.
References
Kimbrell, D. A. & Beutler, B. The evolution and genetics of innate immunity. Nature Rev. Genet. 2, 256–267 (2001)
Elinav, E., Strowig, T., Henao-Mejia, J. & Flavell, R. A. Regulation of the antimicrobial response by NLR proteins. Immunity 34, 665–679 (2011)
Kawai, T. & Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650 (2011)
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001)
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001)
Girardin, S. E. et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 278, 8869–8872 (2003)
Inohara, N. et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J. Biol. Chem. 278, 5509–5512 (2003)
Ogura, Y. et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 276, 4812–4818 (2001)
Kobayashi, K. et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199 (2002)
Collazo, C. M. & Galan, J. E. The invasion-associated type-III protein secretion system in Salmonella–a review. Gene 192, 51–59 (1997)
Isberg, R. R., Voorhis, D. L. & Falkow, S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50, 769–778 (1987)
Beuzón, C. R. et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19, 3235–3249 (2000)
Keestra, A. M. et al. A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. MBio 2, e00266–11 (2011)
Hardt, W. D., Chen, L. M., Schuebel, K. E., Bustelo, X. R. & Galan, J. E. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93, 815–826 (1998)
Daniel, H. & Kottra, G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Eur. J. Phys. 447, 610–618 (2004)
Mellman, I., Fuchs, R. & Helenius, A. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55, 663–700 (1986)
Nakamura, N., Tanaka, S., Teko, Y., Mitsui, K. & Kanazawa, H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J. Biol. Chem. 280, 1561–1572 (2005)
Bonifacino, J. S. & Traub, L. M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 (2003)
Boes, M. et al. T-cell engagement of dendritic cells rapidly rearranges MHC class II transport. Nature 418, 983–988 (2002)
Chow, A., Toomre, D., Garrett, W. & Mellman, I. Dendritic cell maturation triggers retrograde MHC class II transport from lysosomes to the plasma membrane. Nature 418, 988–994 (2002)
Kleijmeer, M. et al. Reorganization of multivesicular bodies regulates MHC class II antigen presentation by dendritic cells. J. Cell Biol. 155, 53–64 (2001)
Chow, A. Y. & Mellman, I. Old lysosomes, new tricks: MHC II dynamics in DCs. Trends Immunol. 26, 72–78 (2005)
Marsh, M., Griffiths, G., Dean, G. E., Mellman, I. & Helenius, A. Three-dimensional structure of endosomes in BHK-21 cells. Proc. Natl Acad. Sci. USA 83, 2899–2903 (1986)
Kufer, T. A., Kremmer, E., Adam, A. C., Philpott, D. J. & Sansonetti, P. J. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell. Microbiol. 10, 477–486 (2008)
Chamaillard, M. et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nature Immunol. 4, 702–707 (2003)
Girardin, S. E. et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300, 1584–1587 (2003)
Barnich, N., Aguirre, J. E., Reinecker, H. C., Xavier, R. & Podolsky, D. K. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-κB activation in muramyl dipeptide recognition. J. Cell Biol. 170, 21–26 (2005)
Rivas, M. A. et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature Genet. 43, 1066–1073 (2011)
Brain, O. et al. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity 39, 521–536 (2013)
Ma, J. K., Platt, M. Y., Eastham-Anderson, J., Shin, J. S. & Mellman, I. MHC class II distribution in dendritic cells and B cells is determined by ubiquitin chain length. Proc. Natl Acad. Sci. USA 109, 8820–8827 (2012)
Shin, J. S. et al. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature 444, 115–118 (2006)
Smed-Sörensen, A. et al. Differential susceptibility to human immunodeficiency virus type 1 infection of myeloid and plasmacytoid dendritic cells. J. Virol. 79, 8861–8869 (2005)
Chan, W. et al. A recombineering based approach for high-throughput conditional knockout targeting vector construction. Nucleic Acids Res. 35, e64 (2007)
Warming, S., Rachel, R. A., Jenkins, N. A. & Copeland, N. G. Zfp423 is required for normal cerebellar development. Mol. Cell. Biol. 26, 6913–6922 (2006)
Liu, H., Sadygov, R. G. & Yates, J. R., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 (2004)
Gilchrist, A. et al. Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 (2006)
Washburn, M. P., Wolters, D. & Yates, J. R., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnol. 19, 242–247 (2001)
Pham, V. C. et al. Complementary proteomic tools for the dissection of apoptotic proteolysis events. J. Proteome Res. 11, 2947–2954 (2012)
Bakalarski, C. E. et al. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 7, 4756–4765 (2008)
Slot, J. W. & Geuze, H. J. Cryosectioning and immunolabeling. Nature Protocols 2, 2480–2491 (2007)
Acknowledgements
The authors thank Mellman laboratory members for discussions, C. Chalouni for excellent advice and assistance with fluorescence microscopy, S. van Dijk and R. Scriwanek for immuno-electron microscopy, and R. Gentleman, V. Dixit and N. Kayagaki for advice. N.N., J.R.L., Q.P., Z.J., C.B., M.S., L.D. and I.M. are full-time employees of Genentech, which funded the research in its entirety.
Author information
Authors and Affiliations
Contributions
N.N., L.D. and I.M. designed the experiments. N.N. performed the experiments. J.R.L. and Q.P. ran mass spectrometry. N.N., Z.J. and C.B. analysed gene expression and mass spectrometry data. A.d.M. and J.K. performed electron microscopy. M.S. generated mouse lines. N.N. and I.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Phagocytosed Salmonella strongly activates NOD2.
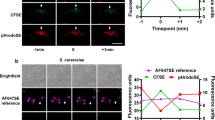
a, NOD2-transfected 293 cells were pre-incubated with or without cytochalasin D, then stimulated with soluble MDP. b, NOD2-transfected 293 cells were stimulated with wild-type or invG mutant of S. typhimurium harbouring a vector or a plasmid carrying Yersinia invasin gene (pInvasin) as in Fig. 1. c, d, 293 cells expressing LAMP1–GFP were incubated with wild-type (c) or sifA mutant (d) of S. typhimurium expressing tagBFP for 1 h, then cultured overnight in the presence of cultured for 24 h in the presence of 20 µg ml−1 gentamycin. The cells were observed under a fluorescence microscope. Bars, 5 µm. e, NOD2- or vector-transfected 293 cells were stimulated with wild-type or sifA mutant of S. typhimurium as in Fig. 1. f, NOD2-transfected 293 cells were stimulated with wild-type or sifA mutant of S. typhimurium harbouring vector or a plasmid carrying SifA gene as in Fig. 1. g, NOD2- or vector-transfected 293 cells were stimulated with wild-type, sopE, sipA, sopE sipA or invG mutant of S. typhimurium. h, NOD2-transfected cells were incubated with rhodamine-conjugated MDP-beads, then uptake by cells was quantified by FACS. One representative result of three independent experiments is shown. Mean of triplicates ± s.d. *P < 0.05 by t-test.
Extended Data Figure 2 LAMP2 and MHCII are enriched in the endo-lysosome fraction of dendritic cells.
a, b, Microscopic images of endo-lysosomes isolated from BMDCs stimulated without (a) or with (b) LPS. BMDCs were cultured with 200-nm diameter iron-oxide particles and ovalbumin (as an endocytic tracer), then incubated without (a) or with 30 ng ml−1 LPS (b) for 3 h. The endo-lysosomes were isolated using magnetic separation from the cells lysed in isotonic buffer. Endocytosed ovalbumin-Alexa555 (OVA), MHCII-GFP, and bright field (BF) are shown. When dendritic cells are stimulated with LPS, isolated endo-lysosomes contained numerous tubular structures (arrows). Bars, 5 µm. c, Total lysate, post-nuclear supernatant (PNS), flow through (unbound), and the fractions bound to column were blotted with antibodies against the marker proteins. Recoveries were calculated based on densitometric analysis using the ImageJ (NIH). LAMP2 and MHCII are enriched 49- and 19-fold, respectively, in fraction 2 with 2% recovery as compared to the cell homogenate. Na+/K+-ATPase (plasma membrane), transferrin receptor (early endosomes), syntaxin-6 (trans-Golgi), and actin (cytosol) are excluded (0.12-, 0.057-, and 0.18-fold enrichment, respectively). d, SILAC mass spectrometry of endo-lysosomal proteins. Abundance ratios of LPS-stimulated endo-lysosomal proteins to unstimulated counterparts are plotted against their relative abundance. 1,436 proteins were identified in the endo-lysosome fractions, 8 and 45 proteins of which were ATP-binding cassette (ABC) transporters and solute carrier family transporters, respectively (shown in red). Eight (ABCA1, ABCA3, TAP1, TAP2, SLC3A2, SLC7A2, SLC7A11 and SLC15A3) were upregulated >twofold by LPS treatment.
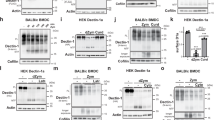
Extended Data Figure 3 Localization of membrane transporters identified in the endo-lysosome fraction.
a, Membrane transporters identified by mass spectrometry were expressed as GFP fusion proteins in 293 cells, then observed by fluorescence microscopy. Endo-lysosomes are visualized with ovalbumin–Alexa555 (OVA) or dextran–Alexa555. Single arrows indicate colocalization of membrane transporters labelled with endocytosed OVA or dextran. Tubular endo-lysosomes were observed only in cells expressing SLC15A3 or SLC15A4 (arrow heads). Double arrows on cells expressing SLC7A11, ABCC4 or ABCC1 mark plasma membrane. b, SLC15A1 (mouse) or SLC15A2 (human) were expressed as GFP fusion proteins in 293 cells, then observed by fluorescence microscopy. Arrows mark plasma membrane. Bars, 5 µm. c, Gene expression of SLC15A1 and SLC15A2 were quantified by RT–PCR. Means ± s.d. (+) denotes LPS-stimulated dendritic cells.
Extended Data Figure 4 Two di-leucine motifs localize mouse SLC15A3 to endo-lysosomes.
a, Di-leucine motifs in mammalian SLC15A3. Mouse and rat proteins have two di-leucine motifs (ExxxLL/V). b, Fluorescence microscopy of di-leucine mutants of mouse SLC15A3–GFP in 293 cells. Alanine substitution of either one in two motifs is insufficient to localize the protein to plasma membrane. Cell surfaces are marked by arrows. Bars, 5 µm. c, 293 cells transfected with an NF-κB-luciferase reporter, NOD2, and either di-leucine mutant of mouse SLC15A3(L19A V20A L288A L289A) or empty vector, were stimulated with soluble MDP for 7 h. Means ± s.d. **P < 0.01.
Extended Data Figure 5 SLC15A3 and SLC15A4 specifically facilitate the signalling induced by MDP-beads.
a, Raw 264.7 cells stably expressing SLC15A3 or SLC15A4 were stimulated with MDP (left) or LPS (right) for 24 h. TNF-α in medium was quantified by ELISA. b, 293 cells transfected with an NF-κB-luciferase reporter, NOD2, and either SLC15A3, SLC15A4 or empty vector, were stimulated with S. typhimurium flagellin or human TNF-α for 7 h. c, SLC15A3-deficient or wild-type BMDCs were stimulated with MDP-beads. IL-1β in medium were quantified by ELISA. d, BMDCs were incubated with MDP–rhodamine-beads, then the uptake was quantified by FACS. e, SLC15A3-deficient or wild-type BMDCs were stimulated with LPS (left) or S. typhimurium (right) for 24 h. IL-6 in medium was quantified by ELISA. Means ± s.d. *P < 0.05 and **P < 0.01.
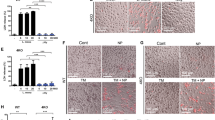
Extended Data Figure 6 Endocytosed MDP are localized to endo-lysosomal tubules tracked along microtubules.
a, 293 cells expressing SLC15A3–GFP were incubated with MDP–rhodamine for 6 h. After replacing with fresh medium without MDP–rhodamine, cells were observed by fluorescence microscopy. b, BMDCs that retrovirally express RFP–tubulin, were incubated with OVA–Alexa488 for 1 h, stimulated with LPS (30 ng ml−1) for 3 h, then observed by fluorescence microscopy. Arrows indicate the endo-lysosomes extending along microtubules. c, MHC-II knock-in BMDCs that retrovirally express SLC15A3–GFP, were incubated with OVA–Alexa647 for 1 h, stimulated with LPS (30 ng ml−1) for 3 h, then observed by fluorescence microscopy. Bars, 5 µm.
Extended Data Figure 7 NOD2 associates to Salmonella-containing phagosomes.
a, 293 cells stably expressing RFP–NOD2 and LAMP1–GFP were incubated with S. typhimurium expressing tagBFP as in Fig. 4a. b, 293 cells expressing RFP–NOD1 were incubated with S. typhimurium expressing EGFP as in Fig. 4a, then observed by fluorescence microscopy. c, Left, 293 cells expressing RFP–NOD2 (FL, full length) or RFP–NOD2-3020insC were incubated with magnetic beads (1 µm), lysed in isotonic buffer, then the phagosomes were isolated using a magnetic column. Right, Phagosomes were isolated from BMDC expressing Flag–NOD2. RFP–NOD2, Flag–NOD2, LAMP1, LAMP2, and GAPDH in the phagosome fractions and total lysates were detected by immunoblotting of duplicated cell culture. NOD2 proteins in phagosomes were calculated by measuring the intensities of protein bands using ImageJ with following formula (1). GAPDH was used to estimate contamination of cytosol in the phagosome fraction. Approximately 20% of NOD2 and 4% of NOD2-3020insC proteins are co-fractionated with LAMP1 or LAMP2 in the phagosome fractions. ([NOD2]fraction/[NOD2]total lysate − [GAPDH]fraction/[GAPDH]total lysate)/([LAMP1]fraction/[LAMP1]total lysate) (1). d, BMDCs retrovirally expressing Flag–NOD2 were incubated with MDP-beads for 24 h, fixed with 4% formaldehyde, permeabilized with 0.05% saponin, then stained with anti-Flag and anti-LAMP2 antibodies, followed by secondary antibodies conjugated with Alexa-dyes. The cells were observed under fluorescence microscopy. MDP-beads in the merged image are shown in blue. BF, bright field. e, 293 cells transfected with an NF-κB-luciferase reporter and either NOD2, RFP–NOD2, or RFP–NOD2-3020insC were incubated with Salmonella as in Fig. 1. The activation of NF-κB was quantified by the luciferase activity. f, Fluorescence microscopy of 293 cells expressing GFP–RIPK2. Endo-lysosomes are labelled with dextran-Alexa647. Bars, 5 µm.
Supplementary information
Supplementary Table 1
This file contains proteins identified in the isolated endo-lysosomes of BMDCs. Relative abundance of proteins (column D, Log2 peak area product) and the ratio of abundance in LPS-stimulated sample to unstimulated sample (column E, Log2 peak area ratio), annotation in UniProt database (column G, Transporter type), intracellular localization by fluorescence microscopy (column H), and gene expression profile by Taqman RT-PCR (column I) are shown. Intracellular localization was examined by expressing GFP-fusion of transporters in 293 cells. The cells were observed under a fluorescence microscope. Gene expression was examined in 11 cell types of human and mouse origin (human: MDC, pDC, HeLa, 293, Caco2, SW480; mouse: BMDC, J774.2, Raw264.7, L929, NIH3T3) by Taqman RT-PCR. Transporters preferentially expressed in DCs and macrophages and upregulated by LPS-stimulation are indicated by closed triangles. The tested transporters are marked by triangles. Previous proteomic analysis of DC-endosomes by Duclos et al. 30 reported 295 proteins including 31 transporters in which out of 30 transporters were found in our dataset of 108 transporters. (XLS 293 kb)
Rights and permissions
About this article
Cite this article
Nakamura, N., Lill, J., Phung, Q. et al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509, 240–244 (2014). https://doi.org/10.1038/nature13133
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13133
This article is cited by
-
Inhibition of S6K lowers age-related inflammation and increases lifespan through the endolysosomal system
Nature Aging (2024)
-
Carnosine regulation of intracellular pH homeostasis promotes lysosome-dependent tumor immunoevasion
Nature Immunology (2024)
-
Chemoproteomic development of SLC15A4 inhibitors with anti-inflammatory activity
Nature Chemical Biology (2024)
-
The NLR gene family: from discovery to present day
Nature Reviews Immunology (2023)
-
In vitro PCR verification that lysozyme inhibits nucleic acid replication and transcription
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.