Abstract
A hexanucleotide repeat expansion (HRE), (GGGGCC)n, in C9orf72 is the most common genetic cause of the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Here we identify a molecular mechanism by which structural polymorphism of the HRE leads to ALS/FTD pathology and defects. The HRE forms DNA and RNA G-quadruplexes with distinct structures and promotes RNA•DNA hybrids (R-loops). The structural polymorphism causes a repeat-length-dependent accumulation of transcripts aborted in the HRE region. These transcribed repeats bind to ribonucleoproteins in a conformation-dependent manner. Specifically, nucleolin, an essential nucleolar protein, preferentially binds the HRE G-quadruplex, and patient cells show evidence of nucleolar stress. Our results demonstrate that distinct C9orf72 HRE structural polymorphism at both DNA and RNA levels initiates molecular cascades leading to ALS/FTD pathologies, and provide the basis for a mechanistic model for repeat-associated neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Richard, G. F., Kerrest, A. & Dujon, B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 72, 686–727 (2008)
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011)
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011)
Kiernan, M. C. et al. Amyotrophic lateral sclerosis. Lancet 377, 942–955 (2011)
Majounie, E. et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11, 323–330 (2012)
Rademakers, R., Neumann, M. & Mackenzie, I. R. Advances in understanding the molecular basis of frontotemporal dementia. Nature Rev. Neurol. 8, 423–434 (2012)
van Langenhove, T., van der Zee, J. & van Broeckhoven, C. The molecular basis of the frontotemporal lobar degeneration–amyotrophic lateral sclerosis spectrum. Ann. Med. 44, 817–828 (2012)
Rollinson, S. et al. Analysis of the hexanucleotide repeat in C9ORF72 in Alzheimer’s disease. Neurobiol. Aging 33, 1846.e5–1846.e6 (2012)
Kohli, M. A. et al. Repeat expansions in the C9ORF72 gene contribute to Alzheimer’s disease in Caucasians. Neurobiol. Aging 34, 1519.e5–1519.e12 (2013)
Majounie, E. et al. Repeat expansion in C9ORF72 in Alzheimer’s disease. N. Engl. J. Med. 366, 283–284 (2012)
Hensman-Moss, D. J. et al. C9orf72 expansions are the most common genetic cause of Huntington disease phenocopies. Neurology 82, 292–299 (2014)
Rutherford, N. J. et al. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiol. Aging 33, 2950.e5–2950.e7 (2012)
Mirkin, S. M. Expandable DNA repeats and human disease. Nature 447, 932–940 (2007)
La Spada, A. R. & Taylor, J. P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature Rev. Genet. 11, 247–258 (2010)
Fratta, P. et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2, 1016 (2012)
Reddy, K., Zamiri, B., Stanley, S. Y. R., Macgregor, R. B. & Pearson, C. E. The Disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 288, 9860–9866 (2013)
Xu, Z. et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl Acad. Sci. USA 110, 7778–7783 (2013)
Mori, K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013)
Ash, P. E. A. et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013)
Donnelly, C. J. et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013)
Lagier-Tourenne, C. et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl Acad. Sci. USA 110, E4530–E4539 (2013)
Sareen, D. et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 5, 208ra149 (2013)
Zu, T. et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl Acad. Sci. USA (2013)
Gellert, M., Lipsett, M. N. & Davies, D. R. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA 48, 2013 (1962)
Huppert, J. L. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 37, 1375 (2008)
Maxam, A. M. & Gilbert, W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA 74, 560–564 (1977)
Kumari, S., Bugaut, A., Huppert, J. L. & Balasubramanian, S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nature Chem. Biol. 3, 218–221 (2007)
Biffi, G., Tannahill, D., McCafferty, J. & Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nature Chem. 5, 182–186 (2013)
Li, H. et al. Ultrasensitive label-free amplified colorimetric detection of p53 based on G-quadruplex MBzymes. Biosens. Bioelectron. 50, 180–185 (2013)
Ong, S.-E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002)
Abdelmohsen, K. & Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 9, 799–808 (2012)
Eulalio, A., Behm-Ansmant, I. & Izaurralde, E. P bodies: at the crossroads of post-transcriptional pathways. Nature Rev. Mol. Cell Biol. 8, 9–22 (2007)
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F.-M. & Lamond, A. I. The nucleolus under stress. Mol. Cell 40, 216–227 (2010)
Hetman, M. & Pietrzak, M. Emerging roles of the neuronal nucleolus. Trends Neurosci. 35, 305–314 (2012)
Hazel, P., Huppert, J., Balasubramanian, S. & Neidle, S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 126, 16405–16415 (2004)
Hellman, L. M. & Fried, M. G. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nature Protocols 2, 1849–1861 (2007)
Sun, D. & Hurley, L. H. in Methods in Molecular Biology (ed. Baumann, P. ) 608, 65–79 (Humana, 2009)
Hsu, R.-J. et al. Long tract of untranslated CAG repeats is deleterious in transgenic mice. PLoS ONE 6, e16417 (2011)
Reddy, K. et al. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 39, 1749–1762 (2011)
Iioka, H., Loiselle, D., Haystead, T. A. & Macara, I. G. Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res. 39, e53 (2011)
Wiśniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nature Methods 6, 359–362 (2009)
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003)
Matunis, M. J., Xing, J. & Dreyfuss, G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 22, 1059–1067 (1994)
Tsoi, H., Lau, T. C.-K., Tsang, S.-Y., Lau, K.-F. & Chan, H. Y. E. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc. Natl Acad. Sci. USA 109, 13428–13433 (2012)
Almeida, S. et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 126, 385–399 (2013)
Uemura, M. et al. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene 31, 1254–1263 (2012)
Acknowledgements
We thank B. J. Traynor for the information on the ALS patient cells, M. DeJesus-Hernandez and R. Rademakers for the genotyping PCR protocols on patient cells, P. J. Tienari for providing additional fibroblast/iPS cell lines, M. J. Matunis for the hnRNP F/H antibody, R. S. Nirujogi for his assistance with the SILAC LC-MS analysis, T. K. Chakravarty for his assistance with transfection of HEK293T cells, the S. Balasubramanian laboratory for generously providing the BG4 plasmid, P. L. Steinwald for purification and assistance in the validation of the BG4 protein, R. M. Powers for assistance in the staining and quantification of P bodies in iPS neurons, and O. Pletnikova for processing human tissues. This work was supported by National Institutes of Health grants NS07432 (J.W.) and NS085207 (R.S. and J.D.R.), the Robert Packard Center for ALS Research at Johns Hopkins and the Muscular Dystrophy Association (J.W.), Target ALS (J.D.R.), the ALS Association (J.W.), the Johns Hopkins Alzheimer’s Disease Research Center NIH P50AG05146 (J.C.T.), National Cancer Institute training grant 5T32CA009110-36 (A.R.H. and E.A.J.S.), Maryland Stem Cell Research Fund (C.J.D.), Judith & Jean Pape Adams Charitable Foundation (R.S.), and the Samuel I. Newhouse Foundation (J.C.T.). We thank P. A. Coulumbe, V. C. Culotta, M. J. Matunis, P. S. Miller, B. A. Learn and members of the J. Wang laboratory for manuscript advice and discussions.
Author information
Authors and Affiliations
Contributions
A.R.H. performed the nucleic acid structure analysis and the in vitro transcription studies. G.P. constructed plasmids for in vitro transcription studies. E.A.J.S. and A.R.H performed and analysed the R-loop assay. A.R.H. performed and analysed the RNA pull-down. M.-S.K., P.G.S. and A.P. performed MS analysis on pull-down fractions. A.R.H. performed all B lymphocyte immunofluorescence and analysis. C.J.D., R.S. and J.D.R. performed all patient-derived iPS motor neuron and human tissue analysis. N.J.M. maintained patient cell cultures. J.D.R. and J.C.T. provided patient tissue samples. A.R.H. and J.W. designed the studies, developed the model, and wrote the paper with insights from G.P. and C.J.D. A.R.H. developed and arranged the figures with insights from J.W. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Spectroscopic characterization of DNA/RNA structural polymorphism.
a, The decanucleotide GGGGCCGGGG from the C9orf72 HRE adopts a parallel G-quadruplex conformation indicated by the CD spectra in the presence of KCl (left). The proposed intermolecular parallel G-quadruplex topology (right) of two decanucleotide sequences that form three stacks of G-quartets 5′ to 3′ with three or four nucleotides, GCCG, forming a loop region. b, The complementary sequence strand for the C9orf72 HRE, (CCCCGG)4, shows no apparent structural differences ± KCl. c, The variability in CD spectra increases with the number of repeats (dotted lines), but the CD spectra can be recapitulated (lines) by fitting a linear combination of three component spectra for hairpin, antiparallel G-quadruplex, and parallel G-quadruplexes from the spectra corresponding to 0 mM KCl (GGGGCC)4, 100 mM KCl (GGGGCC)4, and 100 mM KCl GGGGCCGGGG, respectively. The residuals of the fit are plotted below. d, CD spectral analysis of the thermal stability of the DNA antiparallel G-quadruplex. CD spectra were obtained as described in Methods with temperatures ranging from 25–95 °C in 10 mM Tris-HCl pH 7.4 with 50 mM KCl. This two-state transition was evaluated by singular value decomposition (SVD) of the CD melting spectra (Matlab). The two basis component spectra, shown as left coefficients, represent an antiparallel G-quadruplex and an unfolded single-stranded DNA oligo. The residuals show the difference between the original CD spectrum and the basis component spectra fit. e, Increasing KCl concentrations increase the abundance and stability of antiparallel DNA G-quadruplexes, as shown by ultraviolet–visible melting curves. CD absorbance was normalized to the equation (Abst – min)*(max – min)−1, where Abst is the absorbance at a given temperature, max is the observed maximum absorbance at 295 nm at that KCl concentration, and min is the minimum value obtained for all KCl concentrations. Data were fit to a Boltzmann distribution (Prism) with a lower boundary constraint set to min. f, The decamer HRE sequence forms a parallel G-quadruplex that is less stable than the antiparallel conformation. The data obtained were normalized as described above, except the CD absorptivity was measured at 260 nm. g, CD melting spectrum shows a two-state transition from a folded parallel RNA G-quadruplex state to an unfolded linear state. The CD absorptivity data measured at 260 nm were normalized as described above and fit to a Boltzmann distribution without constraints. h, The (GGGGCC)4 forms a G-quadruplex that is unperturbed by a 3′ biotin label. CD spectra were obtained with and without KCl as in Fig. 1 to identify the stabilization of K+ in the formation of a G-quadruplex that had been chemically modified with the biotin tag. All calculated melting temperatures are provided in Extended Data Table 1 and results are detailed further in Supplementary Results.
Extended Data Figure 2 Depurination of the HRE-containing plasmid template contributes minimally to abortive transcription.
a, Schematic digestion map for analysing depurination levels present on the template plasmid. b, The same patterns are shown for the (CCCCGG)∼70 repeat template and a GFP control template with piperidine treatment and prior acid treatment, confirming that the repeat tract in the repeat-containing template is depurinated basally at nonspecific/spontaneous levels similar to those of the GFP template. The ∼135 nucleotide 5′-end-radiolabelled cleavage fragment in the pCR8-70 samples serves as a size marker for the lower boundary of the repeat region corresponding to the region where the repeats would begin to appear on the gel. More details concerning these findings and the experimental designs are described in Supplementary Results and Methods. c, Densitometric plot profiles of the gel bands from each lane in b are shown to illustrate the depurination results. The left panel shows the small depurination differences between the piperidine-treated template without prior treatment and those that went through the denaturing/annealing treatments with and without KCl. The middle and right panels show that treatment with piperidine causes similar patterns of minimal cleavage for the pCR8-70 and control GFP templates, respectively, as compared to the acid-induced depurination controls, consistent with basal levels of nonspecifically/spontaneously cleaved products. The densitometry analysis was done using ImageJ and plotted in Prism.
Extended Data Figure 3 G-quadruplexes increase abortive transcripts within the C9orf72 HRE region in vitro.
a, A newly developed BG4 nanobody directly binds the K+-dependent G-quadruplex formed by DNA (GGGGCC)4. BG4 was purified from E. coli, and an ELISA experiment using biotinylated DNA (GGGGCC)4, with or without 100 mM KCl during annealing, was used to determine the specificity of BG4, essentially as previously described28. Data show the mean ± s.d. n = 3. b, The ΔCt graph for 100 mM KCl vs 0 mM KCl shows that BG4 pulled down >50-fold more plasmid when the plasmid containing (GGGGCC)21 repeats was annealed in the presence of 100 mM KCl, directly demonstrating the presence of G-quadruplexes formed by the sequence. The DNA pull-down with BG4 is described in the Supplementary Methods. n = 1. c, Formation of G-quadruplexes in the coding region before transcription causes a shift to an earlier termination of abortive transcripts and further loss of the full-length products. The shift is represented below the densitometric plot profile by a significant change in the centre of mass for the repeat region (n = 5). The further loss of full-length transcriptional product is shown on the graph to the right of the transcription profile (n = 3). P values were calculated using a paired fit for each trial assuming a parametric distribution. Data are means ± s.e.m. *P < 0.05; **P < 0.01. d, Accumulation of the abortive transcripts indicates that the repeat-dependent impairment of transcription is not transient but persists over time with and without KCl. Aliquots were removed from the in vitro transcription reactions at different time points and then visualized and analysed as in c.
Extended Data Figure 4 R-loops, and not G-quadruplex formation on nascent RNA transcripts, increase abortive transcription within the C9orf72 HRE region in vitro.
a, RNA transcripts containing many GGGGCC repeats form G-quadruplexes under physiologically relevant KCl concentrations. A colorimetric assay was performed to identify the formation of RNA G-quadruplexes using the enzyme-like peroxidase activity of G-quadruple•haemin complexes29. b, Workflow considerations for the transcriptional assay. The linear plasmid was first annealed ± 100 mM KCl or 100 mM NaCl in 10 mM Tris-HCl, pH 7.4. To prevent salt concentration-dependent effects on the in vitro transcriptional assay, a second adjustment was made to adjust the salts to a final 50 mM concentration in the assay. Reducing the effects on RNA polymerase allowed us to disambiguate the effects of salt on the conformation of DNA versus the nascent RNA. Comparison of DNA annealed in 0 mM KCl and 100 mM NaCl shows similar reduced polymerase processivity, suggesting that possible formation of G-quadruplexes on the nascent RNA transcripts makes a negligible contribution, but does not exclude intrinsic RNA hairpin-induced termination. c, RNase H treatment during transcription reduces the periodic accumulation of abortive transcripts and increases full-length transcripts. Treatment with RNase H, which specifically cleaves RNA•DNA hybrids, during transcription of the C9orf72 repeats causes a shift from truncated transcripts to full-length transcripts, suggestive of the formation of alternative secondary structures, known as R-loops, caused by increased non-duplex DNA during transcription. d, R-loop formation is observed for the plasmid containing the HRE insert, (GGGGCC)21, but not for a GFP insert in a control. Treatment with RNase A removes ssRNA but does not affect R-loops. As shown in Fig. 2c, the HREs induce the formation of R-loops that cause a decrease in the plasmid mobility but can be relieved with RNase H treatment, which specifically cleaves RNA•DNA hybrids. The addition of RNase A and RNase H has little effect on the mobility of the plasmid containing the GFP insert. Radiolabelling the transcriptional products during in vitro transcription confirms the formation of R-loops, demonstrated by the shift consistent with the supercoiled and relaxed plasmids having altered mobility, which is relieved by treatment with both RNases. The transcripts were body-labelled by including 20 µCi of α-[32P]UTP (Perkin Elmer) and then performing the in vitro transcription as previously described. e, There is a significant increase in the R-loop-induced plasmid mobility shift for the (GGGGCC)21 and (GGGGCC)∼70-containing plasmids when compared to the GFP control insert. The shifted supercoiled plasmid bands were quantified by densitometrically measuring each band intensity after treatment with RNase A versus those after treatment with both RNase A and RNase H (ImageJ, NIH). The overlapping densitometric signal of the supercoiled R-loop smear with the circular plasmid band (Fig. 2b) prevented accurate quantification and was excluded. Data are means ± s.e.m. n = 3. *P < 0.05; **P < 0.01.
Extended Data Figure 5 Workflow for the SILAC methodology used for identification and quantification of proteins that bind the hexanucleotide repeats in a structurally dependent manner.
HEK293T cells that had complete incorporation of either heavy-, medium-, or light-labelled isotopes were incubated with either the (GGGGCC)4 G-quadruplex, the (GGGGCC)4 hairpin, or the (CCCCGG)4 hairpin, respectively. The samples were then processed as described in Methods and presented in Fig. 3. The final eluted samples were combined and prepared for trypsin digestion using filter-aided sample preparation (FASP). The buffer was exchanged to remove salts and SDS. The sample was then subject to LC-MS/MS analysis as described in the Methods.
Extended Data Figure 6 Example of mass spectrometry spectra using SILAC obtained from the eluted fractions from the RNA pull-down.
Four sets of spectra representing unique peptide sequences identified for hnRNP U, hnRNP F, RPL7, and hnRNP K show the quantitative RNA structural preferences that can be identified by using the SILAC methodology. In agreement with the work presented in Fig. 3, each protein represented by the unique peptide spectrum here shows an individual preference for either RNA structure dependence, such as NCL (Fig. 3) and hnRNP U, or sequence dependence, as in the cases of hnRNP K, hnRNP F, and RPL7, as shown by the abundance of the different isotope labels provided by the SILAC method. The quantified differences are provided in Supplementary Table 1.
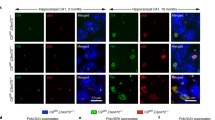
Extended Data Figure 7 Nucleolin defects observed in C9orf72 HRE-linked ALS patients are caused by HRE-containing transcripts.
a, Quantification of nucleolin (NCL) in the nucleus shows significant NCL nuclear dispersion in B lymphocytes, iPS motor neurons, and fibroblasts from C9orf72 HRE patients. A single focal plane was obtained through the centre of the nucleus (Hoechst staining; blue). To quantify the area differences in NCL relative to the size of the nucleus a single lower threshold setting in ImageJ (NIH), ranging from 25–75 (which provided quantification of both dispersed NCL and dense nucleolar NCL), was used to measure the pixel area of NCL relative to the area of the nucleus outlined by the Hoechst staining (depicted in the cartoon). The measurements for the C9orf72 HRE cells were then normalized to those for controls (Prism). Data are means ± s.e.m. n = 110 and n = 112 for 3 cell lines examined for the C9orf72 WT and C9orf72 HRE B lymphocytes, respectively. n = 29 for iPS motor neurons from 2 C9orf72 HRE patients and for the iPS motor neuron controls. A representative fibroblast line for C9orf72 WT, SOD1 D90A ALS, and C9orf72 HRE was quantified. n = 21. *P < 0.05; **P < 0.01; ****P < 0.0001. b, There are no significant changes in NCL protein levels between patients with and without the C9orf72 HRE, despite its mislocalization. The western blotting analysis was performed as described in the Methods, following a typical protocol with the B lymphocyte lysates from patients and controls. The bands were visualized using a quantitative LI-COR imager, and the intensities for NCL and tubulin protein were quantified using Odyssey software. NCL levels were normalized against those of tubulin. Data are means ± s.e.m. n = 6. c, RNA foci are a unique feature of the C9orf72 HRE ALS pathology. Motor cortex tissue from C9orf72 WT (non-ALS) and non-C9orf72 sALS show no RNA foci or NCL colocalization, which is observed in motor cortex tissue from patients carrying the C9orf72 HRE (Fig. 4c). All images were obtained as described in Methods. d, The protein hnRNP F, K, and U, identified from the pull-down (Fig. 3a and Extended Data Fig. 6), show no phenotypic differences in localization between patients carrying the C9orf72 HRE and controls, as previously observed in iPS motor neurons from FTD patients45. n = 3. e, A representative HEK293T cell transfected with the control transcript shows the typical NCL localization (green), in contrast to the nucleolar dispersion observed in cells transfected with (GGGGCC)21 (Fig. 4d). f, NCL is significantly mislocalized when HEK293T cells are transfected with (GGGGCC)21 transcripts, as compared to (GGGGCC)3 or control transcripts. RNA transfections were performed in duplicate, except for No RNA and (GGGGCC)3. Data are means ± s.e.m. n = 27, 50, 37, and 60 for the No RNA, CTL RNA, (GGGGCC)3, and (GGGGCC)21, respectively. ****P < 0.0001. g, There is a significantly decreased cell viability in HEK293T cells transfected with the (GGGGCC)21 abortive RNA transcripts when compared to a control RNA transcript. Cytotoxicity measurements were performed in a 96-well plate with HEK293T cells that had been transfected with 0–500 ng ml−1 of control RNA or (GGGGCC)21 repeat-containing RNAs generated from the in vitro transcription as described earlier and presented in Fig. 2a. Cell viability was measured using the Cytotox Glo kit (Promega), with the fluorescence (viable cells) and luminescence (dead) measured on a Synergy H1 microplate reader. Data are means ± s.d. n = 3. *P < 0.05.
Extended Data Figure 8 Patients carrying the C9orf72 HRE show phenotypes indicative of nucleolar stress.
a, Another nucleolar component, B23 (green), shows a dispersed localization as seen for NCL in B lymphocytes from C9orf72 HRE patients. B23 (mouse monoclonal, B0556, Sigma) immunofluorescence staining followed the manufacturer’s recommendations and the protocol described in Methods. B lymphocytes images were obtained as described in Methods. b, There is a significant increase in the total nuclear area occupied by B23 in B lymphocytes from a patient carrying the C9orf72 HRE when quantified similarly to NCL (Extended Data Fig. 7a). The data are means ± s.e.m. n = 38 and n = 49 for representative C9orf72 WT and C9orf72 HRE B lymphocytes, respectively. *P < 0.05. c, The processing of the 45S pre-rRNA into the mature 28S, 18S, and 5.8S rRNAs that occurs in the nucleolus is depicted. d, Maturation of the 45S pre-rRNA to the 28S, 18S, and 5.8S rRNAs is impaired in patients carrying the C9orf72 HRE. B lymphocytes from patients (n = 6) show a decrease in 45S rRNA processing, but the processing is significantly decreased in the motor cortex tissues of patients carrying the C9orf72 HRE relative to controls (n = 4, 3, and 6 for C9orf72 WT, non-C9orf72 ALS, and C9orf72 HRE, respectively). Furthermore, ALS patients that do not carry the C9orf72 HRE show no significant impairment in 45 s maturation (non-C9orf72 HRE ALS, n = 3). The primers for 45S, 28S, 18S, and 5.8S rRNAs (Extended Data Table 1) were previously described46. The linear-fold change in mature rRNA levels was normalized relative to the parent 45S rRNA levels. Data are means ± s.e.m. *P < 0.05.
Extended Data Figure 9 iPS motor neurons from patients carrying the C9orf72 HRE show an increased number of P bodies and increased sensitivity to tunicamycin.
a, The number of processing bodies (P bodies), but not the size, is significantly increased in iPS motor neurons from patients carrying the C9orf72 HRE. There is an almost twofold increase in the total number of P bodies per iPS neuron from C9orf72 HRE patients. The number and size of P bodies from 30 fields of view were quantified in two independent patient iPS motor neuron lines and two control lines. Data are means ± s.e.m. n = 43. *P < 0.05. b, The representative images of P bodies in iPS neurons were visualized and quantified using a DCP1A antibody (Abnova H00055802-M06, green), and the imaging and analysis are described in Supplementary Methods. c, The percent change in cell death with increasing doses of tunicamycin indicates that it takes lower concentrations of tunicamycin to induce significant toxicity in C9orf72 HRE iPS motor neurons than in control neurons. Moreover, there is a significant difference in tunicamycin responses at 0.3 µM when compared between C9orf72 HRE and C9orf72 WT iPS motor neurons. Each iPS line was differentiated twice and the tunicamycin sensitivity at varying concentrations (0.0, 0.1, 0.3, 0.6, 1.0 and 3.0 µM) was analysed twice. The mean number of propidium iodide (PI)-positive iPS neurons was normalized to that of untreated iPS neurons from the same line. Data are means ± s.e.m. n = 69/58 (0.0), 62/69 (0.1), 75/55 (0.3), 54/49 (0.6), 55/52 (1.0), and 60/53 (3.0) for C9orf72 WT/HRE samples, respectively. **P < 0.01. d, Representative iPS motor neuron images showing the increased PI staining (white) in response to increasing tunicamycin concentrations (Supplementary Methods).
Supplementary information
Supplementary Information
This file contains more technical details of methods, extended analysis of results, additional discussion of the results, and additional references. (PDF 254 kb)
Supplementary Table 1
List of the proteins identified in the SILAC analysis of the protein-RNA interactions. Duplicate LC-MS/MS runs of SILAC samples resulted in a total of 288 protein identifications at a 1% false-discovery rate. The relative abundance of these 288 proteins that bound to the (GGGGCC)4 RNA G-quadruplex (heavy), to the (GGGGCC)4 RNA hairpin (medium), and to the antisense (CCCCGG)4 RNA hairpin (light) is shown in the full list. The proteins with >2 unique peptides and >2-fold change are separately listed. (XLS 116 kb)
Supplementary Table 2
List of patient tissue and cell line information and experimental analyses. (PDF 70 kb)
Rights and permissions
About this article
Cite this article
Haeusler, A., Donnelly, C., Periz, G. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 (2014). https://doi.org/10.1038/nature13124
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13124
This article is cited by
-
Crosstalk between G-quadruplex and ROS
Cell Death & Disease (2023)
-
ALS-linked TDP-43 mutations interfere with the recruitment of RNA recognition motifs to G-quadruplex RNA
Scientific Reports (2023)
-
C9orf72 functions in the nucleus to regulate DNA damage repair
Cell Death & Differentiation (2023)
-
Phenylalanine-tRNA aminoacylation is compromised by ALS/FTD-associated C9orf72 C4G2 repeat RNA
Nature Communications (2023)
-
Mutant GGGGCC RNA prevents YY1 from binding to Fuzzy promoter which stimulates Wnt/β-catenin pathway in C9ALS/FTD
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.