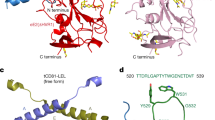
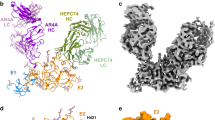
Abstract
Hepatitis C virus (HCV) is a significant public health concern with approximately 160 million people infected worldwide1. HCV infection often results in chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. No vaccine is available and current therapies are effective against some, but not all, genotypes. HCV is an enveloped virus with two surface glycoproteins (E1 and E2). E2 binds to the host cell through interactions with scavenger receptor class B type I (SR-BI) and CD81, and serves as a target for neutralizing antibodies2,3,4. Little is known about the molecular mechanism that mediates cell entry and membrane fusion, although E2 is predicted to be a class II viral fusion protein. Here we describe the structure of the E2 core domain in complex with an antigen-binding fragment (Fab) at 2.4 Å resolution. The E2 core has a compact, globular domain structure, consisting mostly of β-strands and random coil with two small α-helices. The strands are arranged in two, perpendicular sheets (A and B), which are held together by an extensive hydrophobic core and disulphide bonds. Sheet A has an IgG-like fold that is commonly found in viral and cellular proteins, whereas sheet B represents a novel fold. Solution-based studies demonstrate that the full-length E2 ectodomain has a similar globular architecture and does not undergo significant conformational or oligomeric rearrangements on exposure to low pH. Thus, the IgG-like fold is the only feature that E2 shares with class II membrane fusion proteins. These results provide unprecedented insights into HCV entry and will assist in developing an HCV vaccine and new inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lavanchy, D. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 17, 107–115 (2011)
Pileri, P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941 (1998)
Scarselli, E. et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025 (2002)
Sautto, G., Tarr, A. W., Mancini, N. & Clementi, M. Structural and antigenic definition of hepatitis C virus E2 glycoprotein epitopes targeted by monoclonal antibodies. Clin. Dev. Immunol. 2013, 450963 (2013)
Michalak, J. P. et al. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78, 2299–2306 (1997)
Drummer, H. E. & Poumbourios, P. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J. Biol. Chem. 279, 30066–30072 (2004)
Wahid, A. & Dubuisson, J. Virus-neutralizing antibodies to hepatitis C virus. J. Viral Hepat. 20, 369–376 (2013)
Keck, Z. Y. et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 8, e1002653 (2012)
Kong, L. et al. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J. Virol. 86, 13085–13088 (2012)
Kong, L. et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc. Natl Acad. Sci. USA 109, 9499–9504 (2012)
Deng, L. et al. Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc. Natl Acad. Sci. USA 110, 7418–7422 (2013)
Whidby, J. et al. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J. Virol. 83, 11078–11089 (2009)
Krey, T. et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6, e1000762 (2010)
Keck, Z. Y. et al. Analysis of a highly flexible conformational immunogenic domain A in hepatitis C virus E2. J. Virol. 79, 13199–13208 (2005)
Rothwangl, K. B., Manicassamy, B., Uprichard, S. L. & Rong, L. Dissecting the role of putative CD81 binding regions of E2 in mediating HCV entry: putative CD81 binding region 1 is not involved in CD81 binding. Virol. J. 5, 46 (2008)
Lindenbach, B. D., Thiel, H.-J. & Rice, C. M. in Fields Virology (eds Knipe, D. M. & Howley, P. M. ) 1101–1152 (Lippincott Williams & Wilkins, 2007)
White, J. M., Delos, S. E., Brecher, M. & Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43, 189–219 (2008)
Vaney, M. C. & Rey, F. A. Class II enveloped viruses. Cell. Microbiol. 13, 1451–1459 (2011)
El Omari, K., Iourin, O., Harlos, K., Grimes, J. M. & Stuart, D. I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 3, 30–35 (2013)
Li, Y., Wang, J., Kanai, R. & Modis, Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl Acad. Sci. USA 110, 6805–6810 (2013)
Forns, X. et al. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc. Natl Acad. Sci. USA 97, 13318–13323 (2000)
Helle, F. et al. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J. Virol. 84, 11905–11915 (2010)
Lavillette, D. et al. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J. Virol. 81, 8752–8765 (2007)
Li, H. F., Huang, C. H., Ai, L. S., Chuang, C. K. & Chen, S. S. Mutagenesis of the fusion peptide-like domain of hepatitis C virus E1 glycoprotein: involvement in cell fusion and virus entry. J. Biomed. Sci. 16, 89 (2009)
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010)
Kong, L. et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342, 1090–1094 (2013)
Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl Acad. Sci. USA 99, 13419–13424 (2002)
Nielsen, S. S., Moller, M. & Gillilan, R. E. High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J. Appl. Crystallogr. 45, 213–223 (2012)
Petoukhov, M. V. et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 (2012)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Zhang, K. Y., Cowtan, K. & Main, P. Combining constraints for electron-density modification. Methods Enzymol. 277, 53–64 (1997)
Semenyuk, A. V. & Svergun, D. I. GNOM - a program package for small-angle scattering data processing. J. Appl. Crystallogr. 24, 537–540 (1991)
Svergun, D. I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999)
Volkov, V. V. & Svergun, D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003)
Kozin, M. B. & Svergun, D. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 (2001)
Sharma, S. et al. Construct optimization for protein NMR structure analysis using amide hydrogen/deuterium exchange mass spectrometry. Proteins 76, 882–894 (2009)
Tiller, T., Busse, C. E. & Wardemann, H. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods 350, 183–193 (2009)
Mateu, G., Donis, R. O., Wakita, T., Bukh, J. & Grakoui, A. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology 376, 397–407 (2008)
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983)
Katoh, K., Kuma, K., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005)
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009)
Acknowledgements
We acknowledge access to the X25 beamline at NSLS and thank the NSLS staff. NSLS is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. We thank J. Tainer and J. Perry for their support and access to SIBYLS beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory. We thank E. Arnold, J. Bonanno, S. K. Burley, J. Chiu, D. Comoletti, E. Elrod, F. Jiang, S. Khare, P. Lobel, A. Shatkin, A. Stock, J. Shires and A. Thanou for providing helpful comments and assistance. Special thanks to C. M. Rice for providing J6 HCV clone and support. This work was supported by a Yerkes Research Center Base Grant RR-00165 (A.G.) and NIH grants P50 GM103368 (J.M.), R01 AI080659 (J.M.) AI070101 (A.G.) and DK083356 (A.G.). A.G.K. was supported by a grant from the New Jersey Commission on Cancer Research (DFHS13CRP001).
Author information
Authors and Affiliations
Contributions
The project was initiated, designed and supervised by A.G. and J.M. A.G.K., J.W., A.V.Z., A.C. and S.A.Y. designed protein constructs and established purification protocols. A.G.K. prepared all protein crystals. The mouse monoclonal antibody was produced by H.S. and A.G., and was sequenced by C.D.B. and J.J. A.A.P. and A.G. performed the virus neutralization and patient sera ELISA. J.M., M.T.M. and A.G.K. collected, processed and analysed the X-ray crystallographic and SAXS data. A.G.K., J.W. and J.M. wrote the paper with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 eE2, eE2(ΔHVR1) and E2 core are highly soluble and monomeric in solution.
a, A comparison of proteins under reducing and non-reducing conditions is shown by a 10% SDS–PAGE gel with protein standards (Std). b, Size-exclusion chromatography of eE2, eE2(ΔHVR1) and E2 core proteins on a Superdex200 gel filtration column. The elution positions of the void volume (>200 kDa), albumin (66 kDa) and cytochrome C (12.4 kDa) are indicated. Molecular masses of eE2, eE2(ΔHVR1) and E2 core are ∼46 kDa, ∼42 kDa and ∼32 kDa, respectively.
Extended Data Figure 2 eE2 sequence alignment.
Red bent arrows indicate the N- and C-terminal boundaries of the E2 crystallization construct. Cylinders and arrows represent α-helices and β-strands, respectively, and are coloured according to cartoon representation in Fig. 1. CD81 binding regions are bracketed in red; hypervariable regions are bracketed in black. SR-BI binds to HVR1. The asterisks indicate the location of trypsin (blue), chymotrypsin (green) and GluC (magenta) cleavage sites. The binding sites of neutralizing antibodies for which structural information is available are coloured orange for HCV1 and AP33, blue for mAb 8, and purple for HC34-1 and HC34-17.
Extended Data Figure 3 Hydrogen deuterium exchange and limited proteolysis of eE2.
a, The percentage hydrogen deuterium exchange shown at 10, 100 and 1,000 s time points. The secondary structure of E2 core is placed above to emphasize flexible regions. A red arrow indicates the E2 core N terminus. Extra residues (grey) on N and C terminus come from the vector. Potential cleavage sites for trypsin (blue), chymotrypsin (green) and GluC (magenta) are indicated by asterisks. The colour pattern indicates the percentage of exchange. Grey areas are the regions of no coverage. b, Digestion of deglycosylated eE2 with chymotrypsin (left) and GluC (right) reveals a shift from the ∼35-kDa untreated protein (0 min) to ∼25 kDa after digestion. Samples were taken at the indicated time points and analysed by reducing 12% SDS–PAGE gel. Molecular mass protein standards (Std) are indicated. The bands were analysed by N-terminal sequencing and mass spectrometry.
Extended Data Figure 4 Functional analyses of eE2 and E2 core.
a, Antibodies from patient sera infected with HCV genotype 2 show a concentration-dependent binding to eE2 (red) whereas healthy donor sera exhibit only background binding (black). b, Similar binding is observed for E2 core. The measurements were done in triplicate with the error bars representing the standard error of the mean (s.e.m.). c, E2 core (light grey) shows reduced binding to CD81 when compared to eE2 (dark grey) by an ELISA. Bars with stripes indicate E2 binding to a negative control, BSA. The solid black bar indicates CD81 binding to PBS, used to verify the absence of background. The measurements were done in triplicate with the error bars representing the s.e.m. d, eE2 (blue) and CD81-LEL (positive control, grey) inhibit the infection. E2 core (red) shows reduced inhibition. HIV gp140 (black) expressed in the same system was used as a negative control. The measurements were done in triplicate with the error bars representing the s.e.m. e, To rule out the possibility of toxic effects from the recombinant proteins, the cell viability was measured as described in Methods, using similar protein concentrations as in d. f, In an ELISA, 2A12 (red), and an irrelevant antibody, H113 (grey), fail to neutralize HCVcc infection. 2C1 (positive control, black), a mouse monoclonal antibody that binds to the disordered N-terminal region of eE2, blocks infection. The measurements were done in triplicate with the error bars representing the s.e.m.
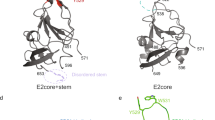
Extended Data Figure 5 E2 core contains an extensive hydrophobic core.
Sheets A and B are held together by an extensive hydrophobic core composed of mostly aromatic amino acids (green) and five disulphide bonds (yellow).
Extended Data Figure 6 eE2 and E2 core do not undergo oligomeric changes at low pH.
a, b, An overlay of E2 core (a) and eE2 (b) elution profiles from Superdex200 gel filtration at pH 7.5 (blue) and pH 5.0 (red). The expected void volume and observed elution positions of individual proteins are indicated. c, The SAXS envelope of CD81-LEL fit with a dimer crystal structure (PDB 1G8Q). The individual proteins of the CD81-LEL dimer are coloured red and blue.
Extended Data Figure 7 Epitope mapping of conformational antibodies on E2 core surface.
a, Surface epitopes of AR1 (orange) are shown. AR1 blocks the E1E2 heterodimer binding to CD81. AR5A (purple) inhibits E1E2 heterodimerization and is mapped on a well-conserved hydrophobic surface of the core. b, Surface of E2 core coloured by electrostatic potential. The view in a and b is identical.
Rights and permissions
About this article
Cite this article
Khan, A., Whidby, J., Miller, M. et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509, 381–384 (2014). https://doi.org/10.1038/nature13117
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13117
This article is cited by
-
Regions of hepatitis C virus E2 required for membrane association
Nature Communications (2023)
-
Entwicklungsansätze für Impfstoffe gegen Hepatitis-C-Virus-Infektionen
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2022)
-
Induction of cross-neutralizing antibodies by a permuted hepatitis C virus glycoprotein nanoparticle vaccine candidate
Nature Communications (2022)
-
Structural insights into hepatitis C virus receptor binding and entry
Nature (2021)
-
Hepatitis C virus vaccine design: focus on the humoral immune response
Journal of Biomedical Science (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.