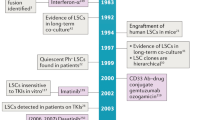
Abstract
In acute myeloid leukaemia (AML), the cell of origin, nature and biological consequences of initiating lesions, and order of subsequent mutations remain poorly understood, as AML is typically diagnosed without observation of a pre-leukaemic phase. Here, highly purified haematopoietic stem cells (HSCs), progenitor and mature cell fractions from the blood of AML patients were found to contain recurrent DNMT3A mutations (DNMT3Amut) at high allele frequency, but without coincident NPM1 mutations (NPM1c) present in AML blasts. DNMT3Amut-bearing HSCs showed a multilineage repopulation advantage over non-mutated HSCs in xenografts, establishing their identity as pre-leukaemic HSCs. Pre-leukaemic HSCs were found in remission samples, indicating that they survive chemotherapy. Therefore DNMT3Amut arises early in AML evolution, probably in HSCs, leading to a clonally expanded pool of pre-leukaemic HSCs from which AML evolves. Our findings provide a paradigm for the detection and treatment of pre-leukaemic clones before the acquisition of additional genetic lesions engenders greater therapeutic resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
16 April 2014
A Correction to this paper has been published: https://doi.org/10.1038/nature13190
References
Fialkow, P. J. et al. Clonal development, stem-cell differentiation, and clinical remissions in acute nonlymphocytic leukemia. N. Engl. J. Med. 317, 468–473 (1987)
McCulloch, E. A., Howatson, A. F., Buick, R. N., Minden, M. D. & Izaguirre, C. A. Acute myeloblastic leukemia considered as a clonal hemopathy. Blood Cells 5, 261–282 (1979)
Vogelstein, B., Fearon, E. R., Hamilton, S. R. & Feinberg, A. P. Use of restriction fragment length polymorphisms to determine the clonal origin of human tumors. Science 227, 642–645 (1985)
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013)
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012)
Yates, L. R. & Campbell, P. J. Evolution of the cancer genome. Nature Rev. Genet. 13, 795–806 (2012)
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011)
Campbell, P. J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010)
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010)
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012)
Notta, F. et al. Evolution of human BCR–ABL1 lymphoblastic leukaemia-initiating cells. Nature 469, 362–367 (2011)
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009)
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010)
Mullighan, C. G. et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322, 1377–1380 (2008)
Shlush, L. I. et al. Cell lineage analysis of acute leukemia relapse uncovers the role of replication-rate heterogeneity and microsatellite instability. Blood 120, 603–612 (2012)
Sgroi, D. C. Preinvasive breast cancer. Annu. Rev. Pathol. 5, 193–221 (2010)
Wistuba, I. I., Mao, L. & Gazdar Smoking molecular damage in bronchial epithelium. Oncogene 21, 7298–7306 (2002)
Balaban, G. B., Herlyn, M., Clark, W. H., Jr & Nowell, P. C. Karyotypic evolution in human malignant melanoma. Cancer Genet. Cytogenet. 19, 113–122 (1986)
Vogelstein, B. et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532 (1988)
Walter, M. J. et al. Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 366, 1090–1098 (2012)
Doulatov, S., Notta, F., Laurenti, E. & Dick, J. E. Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–136 (2012)
Raza, A. & Galili, N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nature Rev. Cancer 12, 849–859 (2012)
Shih, A. H., Abdel-Wahab, O., Patel, J. P. & Levine, R. L. The role of mutations in epigenetic regulators in myeloid malignancies. Nature Rev. Cancer 12, 599–612 (2012)
Busque, L. et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature Genet. 44, 1179–1181 (2012)
Fialkow, P. J., Gartler, S. M. & Yoshida, A. Clonal origin of chronic myelocytic leukemia in man. Proc. Natl Acad. Sci. USA 58, 1468–1471 (1967)
Jan, M. et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 4, 149ra118 (2012)
Miyamoto, T., Weissman, I. L. & Akashi, K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc. Natl Acad. Sci. USA 97, 7521–7526 (2000)
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013)
Yan, X. J. et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature Genet. 43, 309–315 (2011)
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010)
Patel, J. P. et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366, 1079–1089 (2012)
Krönke, J. et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood 122, 100–108 (2013)
Doulatov, S. et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nature Immunol. 11, 585–593 (2010)
Notta, F. et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333, 218–221 (2011)
Laurenti, E. et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nature Immunol. 14, 756–763 (2013)
Kim, H. J. et al. Many multipotential gene-marked progenitor or stem cell clones contribute to hematopoiesis in nonhuman primates. Blood 96, 1–8 (2000)
Fialkow, P. J., Janssen, J. W. & Bartram, C. R. Clonal remissions in acute nonlymphocytic leukemia: evidence for a multistep pathogenesis of the malignancy. Blood 77, 1415–1417 (1991)
Eppert, K. et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature Med. 17, 1086–1093 (2011)
Jankowska, A. M. et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 118, 3932–3941 (2011)
Kikushige, Y. et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell 20, 246–259 (2011)
Walter, M. J. et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25, 1153–1158 (2011)
Chan, S. M. & Majeti, R. Role of DNMT3A, TET2, and IDH1/2 mutations in pre-leukemic stem cells in acute myeloid leukemia. Int. J. Hematol. 98, 648–657 (2013)
Challen, G. A. et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature Genet. 44, 23–31 (2012)
Tadokoro, Y., Ema, H., Okano, M., Li, E. & Nakauchi, H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J. Exp. Med. 204, 715–722 (2007)
Kim, S. J. et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 122, 4086–4089 (2013)
Clappier, E. et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J. Exp. Med. 208, 653–661 (2011)
Inaba, H., Greaves, M. & Mullighan, C. G. Acute lymphoblastic leukaemia. Lancet 381, 1943–1955 (2013)
Yasuda, T. et al. Leukemic evolution of donor-derived cells harboring IDH2 and DNMT3A mutations after allogeneic stem cell transplantation. Leukemia http://dx.doi.org/10.1038/leu.2013.278 (15 October 2013)
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009)
Kottaridis, P. D. et al. Studies of FLT3 mutations in paired presentation and relapse samples from patients with acute myeloid leukemia: implications for the role of FLT3 mutations in leukemogenesis, minimal residual disease detection, and possible therapy with FLT3 inhibitors. Blood 100, 2393–2398 (2002)
Heinrich, V. et al. The allele distribution in next-generation sequencing data sets is accurately described as the result of a stochastic branching process. Nucleic Acids Res. 40, 2426–2431 (2012)
Acknowledgements
We thank all members of the Dick laboratory for critical assessment of this work, A. Khandani, P. Penttilä, N. Simard, T. Velauthapillai and the SickKids-UHN flow facility for technical support, J. Claudio for management of the HALT studies that enabled the genetic analysis described herein, and J. Cui and X.-Z. Yang for curating the human AML samples used in these studies. This work was supported by a Postdoctoral Fellowship Award from the McEwen Centre for Regenerative Medicine with funding made available through the Gentle Ben Charity (L.I.S.), a Canadian Institutes for Health Research (CIHR) fellowship in partnership with the Aplastic Anemia and Myelodysplasia Association of Canada and an award from Vetenskapsradet (S.Z.), and by grants from CIHR, Canadian Cancer Society, Terry Fox Foundation, Genome Canada through the Ontario Genomics Institute, Ontario Institute for Cancer Research with funds from the province of Ontario, a Canada Research Chair, and the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC. This work was also supported by the Cancer Stem Cell Consortium with funding from the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-047), and through the Canadian Institutes of Health Research (CSC-105367). Contributors to the HALT Pan-Leukemia Gene Panel are listed in Supplementary Note 1.
Author information
Authors and Affiliations
Contributions
L.I.S. and S.Z. designed and performed experiments, analysed data and wrote the manuscript; A.M., W.C.C screened AML engraftment in xenotransplantation assays; J.M.B., V.G., J.A.K., A.D.S., A.C.S., K.W.Y., M.D.M. collected AML samples and assembled clinical information; J.A.K. correlated xenotransplantation engraftment data with clinical information; J.L.M., M.D. performed xenotransplantation experiments; J.J.F.M., R.M. performed ddPCR; H.J.K., K.L. performed Sanger sequencing; J.D.M., T.J.H., supervised the targeted sequencing; A.M.K.B. and F.Y. performed and analysed targeted sequencing; Q.M.T., L.D.S. performed DNMT3A data mining. M.D.M. designed the study; J.C.Y.W. supervised AML xenotransplantation screening experiments, designed the study and wrote the manuscript; J.E.D. supervised the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 FLT3-ITD is a late event in patients carrying DNMT3A mutation.
PCR analysis of FLT3-ITD50 in stem/progenitor, mature lymphoid and blast (CD45dim CD33+) cell populations from patient no. 13 (a) and no. 14 (b). FLT3-ITD was present in the blasts from both patients, and also in MLPs from patient no. 14. In contrast, DNMT3Amut without FLT3-ITD was detected in multiple non-blast cell populations (see Extended Data Fig. 2). HSC, haematopoietic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; MLP, multilymphoid progenitor; GMP, granulocyte monocyte progenitor; NK, natural killer cells.
Extended Data Figure 2 Frequent occurrence of DNMT3A mutation without NPM1 mutation in stem/progenitor and mature lymphoid cells in AML patients at diagnosis.
a, Summary of the allele frequency (%) of DNMT3A and NPM1 mutations in stem/progenitor, mature lymphoid, and blast (CD45dim CD33+) cell populations from 11 AML patient peripheral blood samples obtained at diagnosis, as determined by droplet digital PCR (ddPCR). Phenotypically normal cell populations were isolated by fluorescence activated cell sorting according to the strategy depicted in Fig. 2a. Mutant allele frequency ∼50% is consistent with a heterozygous cell population. Departures from 50% mutant allele frequency may be stochastic51, related to clonal heterogeneity, or due to the presence of copy number variations, for example loss of the wild type allele (loss of heterozygosity) or amplification of the mutant allele. NA, no cell population detected; HSC, haematopoietic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte erythroid progenitor; MLP, multilymphoid progenitor; GMP, granulocyte monocyte progenitor; NK, natural killer cells. Blank boxes indicate no DNMT3A or NPM1 mutation detected. b, Representative plots showing ddPCR analysis of DNMT3Amut and NPM1c allele frequency in sorted cell populations from patient no. 11. The mutant allele frequency (%) is indicated on each plot.
Extended Data Figure 3 Phenotypically normal stem/progenitor and mature cell populations are present in AML patient samples at diagnosis, remission and relapse.
Flow cytometric analysis showing the gating strategy used to isolate phenotypically normal stem/progenitor and mature lymphoid cell populations from AML patient samples. Diagnosis and relapse samples are from peripheral blood; remission samples are from bone marrow.
Extended Data Figure 4 Cells bearing mutations in DNMT3A but not NPM1 are present at diagnosis in AML patients and persist at remission and relapse.
Allele frequency of DNMT3A and NPM1 mutations of patients no. 28, 35, 55, and 57 in stem/progenitor, mature and blast (CD45dim CD33+) cell populations, as determined by droplet digital PCR (ddPCR). Cells were isolated from diagnosis (blue), early remission (white), relapse (red) or late remission (yellow) samples. At remission, CD33+ myeloid cells were also analysed. HSC, haematopoietic stem cell; MPP, multipotent progenitor; MLP, multilymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte monocyte progenitor; MEP, megakaryocyte erythroid progenitor; NK, natural killer cells.
Extended Data Figure 5 PreL-HSCs in the peripheral blood of AML patients generate multilineage human grafts in immunodeficient mice.
Summary of results of limiting dilution experiments to assess frequency of pre-leukaemic HSCs generating multilineage grafts after xenotransplantation. Cohorts of NSG mice were transplanted intrafemorally with varying numbers of peripheral blood mononuclear cells from diagnostic samples of AML patient no. 11 (a) and no. 55 (b) and analysed after 8 or 16 weeks by flow cytometry. Engraftment was defined as >0.1% human CD45+ cells in the injected right femur. Shown is the number of mice with multilineage human grafts containing both CD33+ myeloid cells and CD33−CD19+ cells. The frequency of pre-leukaemic HSCs was calculated using the ELDA platform49.
Extended Data Figure 6 Frequent generation of non-leukaemic multilineage human grafts following xenotransplantation of peripheral blood cells from AML patients.
Summary of xenograft characteristics in 123 sublethally irradiated NSG mice transplanted intrafemorally with mononuclear peripheral blood cells from 20 AML patients at diagnosis and analysed after 8 weeks by flow cytometry. The proportion of myeloid (CD33+) and B-lymphoid (CD33−CD19+) cells in the human (CD45+) graft is shown. Leukaemic (AML) engraftment is characterized by a dominant myeloid (CD45dimCD33+) graft, whereas non-leukaemic multilineage grafts contain both lymphoid (predominantly CD33–CD19+ B cells) and myeloid (CD33+) cells. No leukaemic or multilineage graft could be detected in 65/123 mice (53%) in this cohort. Red box indicates AML grafts (27 mice, 22%); blue box indicates multilineage grafts (31 mice, 25%).
Supplementary information
Supplementary Information
This file contains the legends for Supplementary Tables 1-5 (see separate excel files) and Supplementary Notes 1-2. (PDF 163 kb)
Supplementary Table 1
This file contains Supplementary Table 1 (see Supplementary Information file for legend). (XLSX 350 kb)
Supplementary Table 2
This file contains Supplementary Table 2 (see Supplementary Information file for legend). (XLSX 12 kb)
Supplementary Table 3
This file contains Supplementary Table 3 (see Supplementary Information file for legend). (XLSX 14 kb)
Supplementary Table 4
This file contains Supplementary Table 4 (see Supplementary Information file for legend). (XLSX 26 kb)
Supplementary Table 5
This file contains Supplementary Table 5 (see Supplementary Information file for legend). (XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Shlush, L., Zandi, S., Mitchell, A. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014). https://doi.org/10.1038/nature13038
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13038
This article is cited by
-
ATP1A1/BCL2L1 predicts the response of myelomonocytic and monocytic acute myeloid leukemia to cardiac glycosides
Leukemia (2024)
-
Persistent IDH mutations are not associated with increased relapse or death in patients with IDH-mutated acute myeloid leukemia undergoing allogeneic hematopoietic cell transplant with post-transplant cyclophosphamide
Bone Marrow Transplantation (2024)
-
Transcriptome free energy can serve as a dynamic patient-specific biomarker in acute myeloid leukemia
npj Systems Biology and Applications (2024)
-
Crosstalk between DNA methylation and hypoxia in acute myeloid leukaemia
Clinical Epigenetics (2023)
-
FBXO22 promotes leukemogenesis by targeting BACH1 in MLL-rearranged acute myeloid leukemia
Journal of Hematology & Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.