Abstract
Recent clinical trials showed that targeting of inhibitory receptors on T cells induces durable responses in a subset of cancer patients, despite advanced disease. However, the regulatory switches controlling T-cell function in immunosuppressive tumours are not well understood. Here we show that such inhibitory mechanisms can be systematically discovered in the tumour microenvironment. We devised an in vivo pooled short hairpin RNA (shRNA) screen in which shRNAs targeting negative regulators became highly enriched in murine tumours by releasing a block on T-cell proliferation upon tumour antigen recognition. Such shRNAs were identified by deep sequencing of the shRNA cassette from T cells infiltrating tumour or control tissues. One of the target genes was Ppp2r2d, a regulatory subunit of the PP2A phosphatase family. In tumours, Ppp2r2d knockdown inhibited T-cell apoptosis and enhanced T-cell proliferation as well as cytokine production. Key regulators of immune function can therefore be discovered in relevant tissue microenvironments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Galon, J. et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006)
Hamanishi, J. et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl Acad. Sci. USA 104, 3360–3365 (2007)
Mahmoud, S. M. et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 29, 1949–1955 (2011)
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013)
Matsushita, H. et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482, 400–404 (2012)
Oble, D. A., Loewe, R., Yu, P. & Mihm, M. C., Jr Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human melanoma. Cancer Immun. 9, 3 (2009)
DuPage, M., Mazumdar, C., Schmidt, L. M., Cheung, A. F. & Jacks, T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 482, 405–409 (2012)
Schreiber, R. D., Old, L. J. & Smyth, M. J. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570 (2011)
Pagès, F. et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. 27, 5944–5951 (2009)
Rusakiewicz, S. et al. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res. 73, 3499–3510 (2013)
Stumpf, M. et al. Intraepithelial CD8-positive T lymphocytes predict survival for patients with serous stage III ovarian carcinomas: relevance of clonal selection of T lymphocytes. Br. J. Cancer 101, 1513–1521 (2009)
Gabrilovich, D. I. & Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nature Rev. Immunol. 9, 162–174 (2009)
Shiao, S. L., Ganesan, A. P., Rugo, H. S. & Coussens, L. M. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 25, 2559–2572 (2011)
Tanchot, C. et al. Tumor-infiltrating regulatory T cells: phenotype, role, mechanism of expansion in situ and clinical significance. Cancer Microenviron. 6, 147–157 (2013)
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010)
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012)
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012)
Leach, D. R., Krummel, M. F. & Allison, J. P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996)
Wolchok, J. D. et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 369, 122–133 (2013)
Curran, M. A., Montalvo, W., Yagita, H. & Allison, J. P. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc. Natl Acad. Sci. USA 107, 4275–4280 (2010)
Westbrook, T. F. et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848 (2005)
Zender, L. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 (2008)
Luo, B. et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl Acad. Sci. USA 105, 20380–20385 (2008)
Fidler, I. J. Biological behavior of malignant melanoma cells correlated to their survival in vivo . Cancer Res. 35, 218–224 (1975)
Hogquist, K. A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994)
Bellone, M. et al. Relevance of the tumor antigen in the validation of three vaccination strategies for melanoma. J. Immunol. 165, 2651–2656 (2000)
Overwijk, W. W. et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J. Exp. Med. 198, 569–580 (2003)
Paolino, M. & Penninger, J. M. Cbl-b in T-cell activation. Semin. Immunopathol. 32, 137–148 (2010)
Zheng, Y., Zha, Y. & Gajewski, T. F. Molecular regulation of T-cell anergy. EMBO Rep. 9, 50–55 (2008)
Doody, K. M., Bourdeau, A. & Tremblay, M. L. T-cell protein tyrosine phosphatase is a key regulator in immune cell signaling: lessons from the knockout mouse model and implications in human disease. Immunol. Rev. 228, 325–341 (2009)
Tamiya, T., Kashiwagi, I., Takahashi, R., Yasukawa, H. & Yoshimura, A. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler. Thromb. Vasc. Biol. 31, 980–985 (2011)
Barr, F. A., Elliott, P. R. & Gruneberg, U. Protein phosphatases and the regulation of mitosis. J. Cell Sci. 124, 2323–2334 (2011)
Muranski, P. et al. Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood 112, 362–373 (2008)
Koller, B. H., Marrack, P., Kappler, J. W. & Smithies, O. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science 248, 1227–1230 (1990)
Torres, A. J., Contento, R. L., Gordo, S., Wucherpfennig, K. W. & Love, J. C. Functional single-cell analysis of T-cell activation by supported lipid bilayer-tethered ligands on arrays of nanowells. Lab Chip 13, 90–99 (2013)
Han, Q., Bradshaw, E. M., Nilsson, B., Hafler, D. A. & Love, J. C. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip 10, 1391–1400 (2010)
Mochida, S., Maslen, S. L., Skehel, M. & Hunt, T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330, 1670–1673 (2010)
Chiang, C. W. et al. Protein phosphatase 2A dephosphorylation of phosphoserine 112 plays the gatekeeper role for BAD-mediated apoptosis. Mol. Cell. Biol. 23, 6350–6362 (2003)
Chuang, E. et al. The CD28 and CTLA-4 receptors associate with the serine/threonine phosphatase PP2A. Immunity 13, 313–322 (2000)
Eitelhuber, A. C. et al. Dephosphorylation of Carma1 by PP2A negatively regulates T-cell activation. EMBO J. 30, 594–605 (2011)
Tao, J. et al. JNK2 negatively regulates CD8+ T cell effector function and anti-tumor immune response. Eur. J. Immunol. 37, 818–829 (2007)
Johnson, L. A. et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114, 535–546 (2009)
Brenner, M. K. & Heslop, H. E. Adoptive T cell therapy of cancer. Curr. Opin. Immunol. 22, 251–257 (2010)
Turtle, C. J., Hudecek, M., Jensen, M. C. & Riddell, S. R. Engineered T cells for anti-cancer therapy. Curr. Opin. Immunol. 24, 633–639 (2012)
Kalos, M. & June, C. H. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity 39, 49–60 (2013)
Restifo, N. P., Dudley, M. E. & Rosenberg, S. A. Adoptive immunotherapy for cancer: harnessing the T cell response. Nature Rev. Immunol. 12, 269–281 (2012)
Ashton, J. M. et al. Gene sets identified with oncogene cooperativity analysis regulate in vivo growth and survival of leukemia stem cells. Cell Stem Cell 11, 359–372 (2012)
Gerber, S. A., Rush, J., Stemman, O., Kirschner, M. W. & Gygi, S. P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl Acad. Sci. USA 100, 6940–6945 (2003)
Acknowledgements
This work was supported by the National Institutes of Health (Transformative Research Award 1R01CA173750 to K.W.W.), the Melanoma Research Alliance (to K.W.W.), the DF/HCC–MIT Bridge Project and the Lustgarten Foundation (to K.W.W., J.C.L. and H.L.P.), Novartis Institutes of Biomedical Research (to K.W.W.), the Koch Institute Support Grant P30-CA14051 from the National Cancer Institute, the American Cancer Society John W. Thatcher, Jr Postdoctoral Fellowship in Melanoma Research (to D.R.S.), the Terri Brodeur Breast Cancer Foundation Postdoctoral Fellowship (to P.Z.) and a NIH T32 grant (AI07386 to D.A.A.A.).
Author information
Authors and Affiliations
Contributions
K.W.W., P.Z., S.J.T., G.D. and H.C. contributed to the overall study design; K.W.W., P.Z. and D.R.S. designed experiments; P.Z., D.A.A.A. and H.C. developed procedure for lentiviral infection of T cells and optimized approaches for adoptive T-cell therapy; P.Z., D.R.S. and D.A.A.A. performed shRNA screen; G.S.C., D.E.R. and N.H. provided pooled shRNA library and advice on shRNA screen; Y.N. and G.D. provided B16-Ova cell line and advice on tumour model; A.J.T. and J.C.L. performed nano-well analysis of cytokine production, V.C. and S.J.T. performed histological studies, W.P. performed protein quantification by mass spectrometry, S.K.D. and H.L.P. provided mouse models; J.B., K.E. and J.L. performed microarray analysis; K.W.W., P.Z. and D.R.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
K.W.W. and G.D. served as consultants to Novartis.
Additional information
The access number for microarray data is GSE53388 in the Genomic Spatial Event (GSE) database.
Extended data figures and tables
Extended Data Figure 1 In vivo RNAi screening procedure.
a, Infection of CD8+ T cells from Rag1−/−/OT-I TCR transgenic mice with shRNA pools. T cells were either activated with anti-CD3/CD28 beads or exposed to recombinant murine IL-7/IL-15 for 48 h. T cells were then infected with a LacZ control shRNA lentiviral vector and cultured for an additional three days. Transduction efficiency was determined based on expression of the Thy1.1 reporter encoded by the lentiviral vector. Cytokine-cultured T cells expressing the LacZ control shRNA were then stained with a panel of activation markers (blue lines; isotype control, shaded). The majority of infected T cells showed a central memory phenotype (CD62L+CD44+). b, Representative flow cytometry plots of OT-I T cells sorted from tumours and secondary lymphoid organs for deep sequencing analysis (dLN, tumour-draining lymph node; irLN, irrelevant lymph node). CD8+Vα2+Vβ5+Thy1.1+ cells were sorted and genomic DNA was extracted for PCR amplification of the shRNA cassette. c, Deep sequencing results from T-cell dysfunction screen. shRNA sequencing reads for genes positive in secondary screen are plotted in comparison to spleen for tumours (red), irrelevant lymph nodes (irLN, blue) and tumour-draining lymph nodes (dLN, green), with dashed lines indicating a deviation of log2 from the diagonal. Data show enrichment of particular shRNAs representing these genes in tumours compared to spleens or lymph nodes. d, Deep sequencing results from kinase and phosphatase screen, as described in c.
Extended Data Figure 2 Validation of shRNAs from in vivo RNAi screen.
a, FACS-based analysis of T-cell enrichment in tumours. Positive shRNAs from deep sequencing analysis were cloned into vectors driving expression of one of four distinct fluorescent proteins (TFP, GFP, RFP, ametrine) or Thy1.1. OT-I T cells were transduced with shRNA vectors and the five populations of T cells (normalized for transduction efficiency) were co-injected into B16-Ova tumour-bearing mice. T cells were isolated from tumours and spleens on day 7, and the percentage of reporter-positive CD8+Vα2+Vβ5+ T cells was determined by flow cytometry. b, FACS analysis of T-cell enrichment in tumours compared to spleen (as described above) for cells expressing a panel of Ppp2r2d or Cblb shRNAs (upper panels). Also, Ppp2r2d and Cblb mRNA levels were measured by qPCR before T-cell transfer (lower panels). The strongest T-cell enrichment in tumours was observed for shRNAs with >80% knockdown efficiency at the mRNA level (shRNAs 1 and 2 for both Ppp2r2d and Cblb). Data represent biological replicates (n = 3), each value represents mean ± s.d.
Extended Data Figure 3 Specificity of Ppp2r2d shRNA.
a, Generation of mutant Ppp2r2d cDNA with wild-type protein sequence but disrupted shRNA binding site. Both mutant and wild-type Ppp2r2d cDNAs were cloned into a modified pLKO.3G vector with a 2A peptide ribosomal skip sequence and GFP. This approach resulted in stoichiometric expression of Ppp2r2d protein and GFP in EL4 thymoma cells. GFP-expressing EL4 cells were sorted to purity and transduced with LacZ or Ppp2r2d shRNA vectors expressing a Thy1.1 reporter. shRNA-transduced (Thy1.1+) cells were analysed by flow cytometry for GFP expression. The Ppp2r2d shRNA reduced GFP levels when wild-type Ppp2r2d cDNA, but not when mutant Ppp2r2d cDNA was co-expressed. b, Expression of Ppp2r2d mutant cDNA prevents phenotype induced by Ppp2r2d shRNA. OT-I T cells were transduced with a vector encoding LacZ shRNA, Ppp2r2d shRNA or Ppp2r2d shRNA plus mutant Ppp2r2d cDNA. The different T-cell populations were normalized for transduction efficiency and co-injected into B16-Ova tumour-bearing mice. The percentage of each T-cell population in tumours and spleens was quantified by gating on CD8+Vα2+Vβ5+ T cells; transduced cells were detected based on expression of Thy1.1 or ametrine/GFP fluorescent reporters (representative data from 2 independent experiments, n = 3 mice per experiment). c, qPCR analysis for Ppp2r2d expression in OT-I T cells transduced with LacZ shRNA, Ppp2r2d shRNA, and Ppp2r2d shRNA plus Ppp2r2d mutant cDNA. Data represent biological replicates (n = 3), each value represents mean ± s.d.
Extended Data Figure 4 Expression profiles of gene-silenced CD8 T cells in tumours.
OT-I T cells were transduced with lentiviral vectors driving expression of one of five experimental shRNAs or LacZ control shRNA. T cells were injected into day 14 B16-Ova tumour-bearing mice and isolated from tumours and spleens 7 days later. Cells were sorted to high purity and total RNA was obtained for Affymetrix gene expression profiling. For each shRNA, arrays were performed in triplicate (6 mice per group). a, Two genes (Egr2 and Ptpn2) have known functions in T cells. Enrichment in tumour versus spleen was calculated based on deep sequencing results from the secondary screen. b, Clustering of mean expression levels for mRNAs found to be significantly regulated by T cells in spleens or tumours expressing the LacZ control shRNA or one of five experimental shRNAs. Significant expression differences were defined as an ANOVA P value ≤ 0.01 between T cells expressing LacZ control shRNA or one of five experimental shRNAs (Alk, Arhgap5, Egr2, Ptpn2 or Ppp2r2d) (JMP-Genomics 6.0, SAS Institute). mRNAs significantly regulated in one or more treatment groups are shown after clustering (fast Ward). c, Venn diagram showing overlaps between expression signatures by tumour-infiltrating T cells transduced with one of the five experimental shRNAs (signatures defined as an ANOVA P ≤ 0.01 as described above). Indicated are the numbers of overlapping probe IDs for any combination of the 5 signatures, as indicated by the overlapping ovals. The significance of the overlaps versus those expected by random chance (Fisher’s exact test) is shown in the accompanying table.
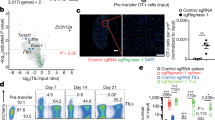
Extended Data Figure 5 Ppp2r2d shRNA enhances T-cell proliferation and reduces apoptosis.
a, Proliferation of Ppp2r2d shRNA-expressing T cells in tumours and tumour-draining lymph nodes. OT-I T cells expressing Ppp2r2d or LacZ shRNAs were labelled with CFSE and injected into B16-Ova tumour-bearing mice. T cells were isolated from the indicated organs on days 1, 3, 5 and 7 to examine the extent of T-cell proliferation based on CFSE dilution. T cells that had not diluted CFSE (non-dividing cells) were quantified (right). b, Viability of tumour-infiltrating T cells. OT-I T cells expressing Ppp2r2d or LacZ shRNAs were injected into B16-Ova tumour-bearing mice. T cells were isolated on day 7 and apoptosis was assessed by intracellular staining with an antibody specific for activated caspase-3 (some T-cell death may have been caused by the isolation procedure from tumours). c, Intracellular cytokine staining for IFN-γ by LacZ and Ppp2r2d shRNA-expressing T cells collected from B16-Ova tumours (primary flow cytometry analysis for data summarized in Fig. 4d); T cells were labelled with CFSE before injection. Data for all experiments are representative of two independent trials. Statistical analysis was performed on biological replicates (n = 3); *P ≤ 0.05, **P ≤ 0.01, two-sided Student’s t-test. Each value represents mean ± s.d.
Extended Data Figure 6 Phenotypic characterization using memory, activation and exhaustion markers.
a, The majority of adoptively transferred OT-I cells have a memory phenotype in lymph nodes but an effector phenotype in tumours. Cytokine pre-treated cells expressing Ppp2r2d or LacZ shRNAs were injected into mice bearing day 14 B16-Ova tumours. On day 7 following transfer, T cells were collected from the indicated organs and stained with CD62L and CD44 antibodies. FACS analysis of shRNA-expressing OT-I cells was performed by gating on CD8/Thy1.1 double-positive cells. b, Analysis of exhaustion markers. OT-I cells were collected from draining lymph nodes and tumours of mice and stained with antibodies specific for TIM-3, LAG-3, PD-1 and CD25. For all experiments (n = 3 biological replicates; *P ≤ 0.05, **P ≤ 0.01, two-sided Student’s t-test); each value represents mean ± s.d.
Extended Data Figure 7 Mechanisms of anti-tumour activity of Ppp2r2d-silenced T cells.
a, Intracellular staining for granzyme B by OT-I T cells in tumour-draining lymph nodes and tumours. b, Infiltration of shRNA-expressing T cells into tumours. OT-I T cells were transduced with LacZ or Ppp2r2d shRNA vectors encoding a GFP reporter and injected into B16-Ova tumour-bearing mice. After 7 days, tumours were excised and frozen sections stained with anti-GFP and DAPI to enumerate shRNA-expressing OT-I T cells in tumours. c, Tumour cell apoptosis. TUNEL immunohistochemistry was performed on tissue sections and apoptotic cells were quantified. d, MHC class I expression by tumour cells. Tumours were digested with collagenase and stained with CD45.2 and H-2Kb antibodies. FACS analysis for H-2Kb expression was performed by gating on CD45.2-negative melanoma cells. Data represent biological replicates (n = 3), each value represents mean ± s.d.
Supplementary information
Supplementary Table 1
This file contains the results from primary and secondary screens. It lists gene name, gene ID, shRNA clone ID, targeting sequence and enrichment of shRNAs in tumors relative to spleen. The first tab shows results from primary screens, the second tab data from secondary screens. (XLS 1303 kb)
Rights and permissions
About this article
Cite this article
Zhou, P., Shaffer, D., Alvarez Arias, D. et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 506, 52–57 (2014). https://doi.org/10.1038/nature12988
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12988
This article is cited by
-
Targeting protein phosphatases in cancer immunotherapy and autoimmune disorders
Nature Reviews Drug Discovery (2023)
-
KCNAB2 overexpression inhibits human non-small-cell lung cancer cell growth in vitro and in vivo
Cell Death Discovery (2023)
-
A T cell resilience model associated with response to immunotherapy in multiple tumor types
Nature Medicine (2022)
-
Cannabis suppresses antitumor immunity by inhibiting JAK/STAT signaling in T cells through CNR2
Signal Transduction and Targeted Therapy (2022)
-
CD8+ T cell differentiation and dysfunction in cancer
Nature Reviews Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.