Abstract
Ancient genomic sequences have started to reveal the origin and the demographic impact of farmers from the Neolithic period spreading into Europe1,2,3. The adoption of farming, stock breeding and sedentary societies during the Neolithic may have resulted in adaptive changes in genes associated with immunity and diet4. However, the limited data available from earlier hunter-gatherers preclude an understanding of the selective processes associated with this crucial transition to agriculture in recent human evolution. Here we sequence an approximately 7,000-year-old Mesolithic skeleton discovered at the La Braña-Arintero site in León, Spain, to retrieve a complete pre-agricultural European human genome. Analysis of this genome in the context of other ancient samples suggests the existence of a common ancient genomic signature across western and central Eurasia from the Upper Paleolithic to the Mesolithic. The La Braña individual carries ancestral alleles in several skin pigmentation genes, suggesting that the light skin of modern Europeans was not yet ubiquitous in Mesolithic times. Moreover, we provide evidence that a significant number of derived, putatively adaptive variants associated with pathogen resistance in modern Europeans were already present in this hunter-gatherer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Accessions
Sequence Read Archive
Data deposits
Alignment data are available through the Sequence Read Archive (SRA) under accession numbers PRJNA230689 and SRP033596.
References
Keller, A. et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nature Commun. 3, 698 (2012)
Sánchez-Quinto, F. et al. Genomic affinities of two 7,000-year-old Iberian hunter-gatherers. Curr. Biol. 22, 1494–1499 (2012)
Skoglund, P. et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336, 466–469 (2012)
Laland, K. N., Odling-Smee, J. & Myles, S. How culture shaped the human genome: bringing genetics and the human sciences together. Nature Rev. Genet. 11, 137–148 (2010)
Rasmussen, M. et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463, 757–762 (2010)
Rasmussen, M. et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science 334, 94–98 (2011)
Vidal Encinas, J. M. & Prada Marcos, M. E. Los hombres mesolíticos de La Braña-Arintero (Valdelugueros, León) (León: Junta de Castilla y León, 2010)
Overballe-Petersen, S., Orlando, L. & Willerslev, E. Next-generation sequencing offers new insights into DNA degradation. Trends Biotechnol. 30, 364–368 (2012)
Allentoft, M. E. et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. R. Soc. B Biol. Sci. 279, 4824–4733 (2012)
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006)
Nelson, M. R. et al. The population reference sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 83, 347–358 (2008)
Novembre, J. et al. Genes mirror geography within Europe. Nature 456, 98–101 (2008)
An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Surakka, I. et al. Founder population-specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging. Genome Res. 20, 1344–1351 (2010)
Raghavan, M. et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014)
Reich, D., Thangaraj, K., Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009)
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010)
Perry, G. H. et al. Diet and the evolution of human amylase gene copy number variation. Nature Genet. 39, 1256–1260 (2007)
Grossman, S. R. et al. Identifying recent adaptations in large-scale genomic data. Cell 152, 703–713 (2013)
Lamason, R. L. et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786 (2005)
Norton, H. L. et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol. Biol. Evol. 24, 710–722 (2007)
Sturm, R. A. & Duffy, D. L. Human pigmentation genes under environmental selection. Genome Biol. 13, 248 (2012)
Sturm, R. A. et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 82, 424–431 (2008)
Walsh, S. et al. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci. Int. Genet. 7, 98–115 (2013)
Aoshi, T., Koyama, S., Kobiyama, K., Akira, S. & Ishii, K. J. Innate and adaptive immune responses to viral infection and vaccination. Curr. Opin. Virol. 1, 226–232 (2011)
Moresco, E. M. Y., LaVine, D. & Beutler, B. Toll-like receptors. Curr. Biol. 21, R488–R493 (2011)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Huson, D. H., Mitra, S., Ruscheweyh, H.-J., Weber, N. & Schuster, S. C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 21, 1552–1560 (2011)
Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F. & Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013)
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010)
Acknowledgements
The authors thank L. A. Grau Lobo (Museo de León) for access to the La Braña specimen, M. Rasmussen and H. Schroeder for valid input into the experimental work, and M. Raghavan for early access to Mal'ta genome data. Sequencing was performed at the Danish National High-Throughput DNA-Sequencing Centre, University of Copenhagen. The POPRES data were obtained from dbGaP (accession number 2038). The authors are grateful for financial support from the Danish National Research Foundation, ERC Starting Grant (260372) to TM-B, and (310372) to M.G.N., FEDER and Spanish Government Grants BFU2012-38236, the Spanish Multiple Sclerosis Netowrk (REEM) of the Instituto de Salud Carlos III (RD12/0032/0011) to A.N., BFU2011-28549 to T.M.-B., BFU2012-34157 to C.L.-F., ERC (Marie Curie Actions 300554) to M.E.A., NIH NRSA postdoctoral fellowship (F32GM106656) to C.W.K.C., NIH (R01-HG007089) to J.N., NSF postdoctoral fellowship (DBI-1103639) to M.D., the Australian NHMRC to R.A.S. and a predoctoral fellowship from the Basque Government (DEUI) to I.O.
Author information
Authors and Affiliations
Contributions
C.L.-F. and E.W. conceived and lead the project. M.E.P. and J.M.V.E. provided anthropological and archaeological information. O.R. and M.E.A. performed the ancient extractions and library construction, respectively. I.O., M.E.A., F.S.-Q., J.P.-M., S.R., O.R., M.F.-C. and T.M.-B. performed mapping, SNP calling, mtDNA assembly, contamination estimates and different genomic analyses on the ancient genome. I.O., F.S.-Q., G.S., C.W.K.C., M.D., J.A.R., J.Q., O.R., U.M.M. and A.N. performed functional, ancestry and population genetic analyses. R.N. and J.N. coordinated the ancestry analyses. M.G.N., R.A.S. and P.S. coordinated the immunological, pigmentation and selection analyses, respectively. I.O., M.E.A., T.M.-B., E.W. and C.L.-F. wrote the majority of the manuscript with critical input from R.N., M.G.N., J.N., R.A.S., P.S. and A.N.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Alignment and coverage statistics of the La Braña 1 genome.
a, Alignment summary of the La Braña 1 sequence data to hg19 assembly. b, Coverage statistics per chromosome. The percentage of the chromosome covered by at least one read is shown, as well as the mean read depth of all positions and positions covered by at least one read. c, Percentage of the genome covered at different minimum read depths.
Extended Data Figure 2 Damage pattern of La Braña 1 sequenced reads.
a, b, Frequencies of C to T (red) and G to A (blue) misincorporations at the 5′ end (left) and 3′ end (right) are shown for the nuclear DNA (nuDNA) (a) and mtDNA (b). c, d, Fragment length distribution of reads mapping to the nuclear genome (c) and mtDNA genome (d). Coefficients of determination (R2) for an exponential decline are provided for the four different data sets. The exponential coefficients for the four data sets correspond to the damage fraction (λ); e is the base of the natural logarithm.
Extended Data Figure 3 Genetic affinities of the La Braña 1 genome.
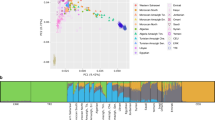
a, PCA of the La Braña 1 SNP data and the 1000 Genomes Project European individuals. b, PCA of La Braña 1 versus world-wide data genotyped with the Illumina Omni 2.5M array. Continental terms make reference to each Omni population grouping as follows: Africans, Yoruba and Luyha; Asians, Chinese (Beijing, Denver, South, Dai), Japanese and Vietnamese; Europeans, Iberians, Tuscans, British, Finns and CEU; and Indian Gujarati from Texas. c, Each panel shows PC1 and PC2 based on the PCA of one of the ancient samples with the merged POPRES+FINHM sample, before Procrustes transformation. The ancient samples include the La Braña 1 sample and four Neolithic samples from refs 1 and 3.
Extended Data Figure 4 Allele-sharing analysis.
Each panel shows the allele-sharing of a particular Neolithic sample from refs 1 and 3 with La Braña 1 sample. The sample IDs are presented in the upper left of each panel (Ajv52, Ajv70, Ire8, Gok4 and Ötzi). In the upper right of each panel, the Pearson’s correlation coefficient is given with the associated P value.
Extended Data Figure 5 Pairwise outgroup f3 statistics.
a, Sardinian versus Karitiana. b, Sardinian versus Han. c, La Braña 1 versus Mal’ta. d, Sardinian versus Mal’ta. e, La Braña 1 versus Karitiana. The solid line represents y = x.
Extended Data Figure 6 Analysis of heterozygosity.
a, Heterozygosity distributions of La Braña 1 and modern individuals with similar coverage from the 1000 Genomes Project (using 1-Mb windows with 200 kb overlap). CEU, northern- and western-European ancestry. CHB, Han Chinese; FIN, Finns; GBR, Great Britain; IBS, Iberians; JPT, Japanese; LWK, Luhya; TSI, Tuscans; YRI, Yorubans. b, Heterozygosity values in 1-Mb windows (with 200 kb overlap) across each chromosome.
Extended Data Figure 7 Amylase copy-number analysis.
a, Size distribution of diploid control regions. b, AMY1 gene copy number in La Braña 1. CN, copy number; DGV, Database of Genomic Variation. c, La Braña 1 AMY1 gene copy number in the context of low- and high-starch diet populations. d, Classification of low- and high-starch diet individuals based on AMY1 copy number. Using data from ref. 18, individuals were classified as in low-starch (less or equal than) or high-starch (higher than) categories and the fraction of correct predictions was calculated. In addition, we calculated the random expectation and 95% limit of low-starch-diet individuals classified correctly at each threshold value.
Extended Data Figure 8 Neighbouring variants for three diagnostic SNPs related to immunity.
a, rs2745098 (PTX4 gene). b, rs11755393 (UHRF1BP1 gene). c, rs10421769 (GPATCH1 gene). For PTX4, UHRF1BP1 and GPATCH1, La Braña 1 displays the derived allele and the European-specific haplotype, indicating that the positive-selection event was already present in the Mesolithic. Blue, ancestral; red, derived.
Extended Data Figure 9 Metagenomic analysis of the non-human reads.
a, Domain attribution of the reads that did not map to hg19. b, Proportion of different Bacteria groups. c, Proportion of different types of Proteobacteria. d, Microbial attributes of the microbes present in the La Braña 1 sample.
Supplementary information
Supplementary Information
This file contains Supplementary Text, additional references and Supplementary Tables 1-26 (see Contents for more details). (PDF 3741 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Olalde, I., Allentoft, M., Sánchez-Quinto, F. et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228 (2014). https://doi.org/10.1038/nature12960
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12960
This article is cited by
-
The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes
Scientific Data (2024)
-
Ancient DNA suggests anaemia and low bone mineral density as the cause for porotic hyperostosis in ancient individuals
Scientific Reports (2023)
-
Genetic continuity, isolation, and gene flow in Stone Age Central and Eastern Europe
Communications Biology (2023)
-
Bioarchaeological and palaeogenomic portrait of two Pompeians that died during the eruption of Vesuvius in 79 AD
Scientific Reports (2022)
-
Is there still evolution in the human population?
Biologia Futura (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.