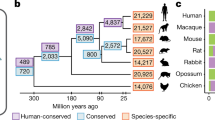
Abstract
Only a very small fraction of long noncoding RNAs (lncRNAs) are well characterized. The evolutionary history of lncRNAs can provide insights into their functionality, but the absence of lncRNA annotations in non-model organisms has precluded comparative analyses. Here we present a large-scale evolutionary study of lncRNA repertoires and expression patterns, in 11 tetrapod species. We identify approximately 11,000 primate-specific lncRNAs and 2,500 highly conserved lncRNAs, including approximately 400 genes that are likely to have originated more than 300 million years ago. We find that lncRNAs, in particular ancient ones, are in general actively regulated and may function predominantly in embryonic development. Most lncRNAs evolve rapidly in terms of sequence and expression levels, but tissue specificities are often conserved. We compared expression patterns of homologous lncRNA and protein-coding families across tetrapods to reconstruct an evolutionarily conserved co-expression network. This network suggests potential functions for lncRNAs in fundamental processes such as spermatogenesis and synaptic transmission, but also in more specific mechanisms such as placenta development through microRNA production.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
Gene Expression Omnibus
Sequence Read Archive
Data deposits
The sequencing data have been deposited in the Gene Expression Omnibus (accession GSE43520) and SRA (PRJNA186438 and PRJNA202404).
References
Kosiol, C. et al. Patterns of positive selection in six mammalian genomes. PLoS Genet. 4, e1000144 (2008)
Brawand, D. et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011)
Khalil, A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11667–11672 (2009)
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011)
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012)
Guttman, M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009)
Guttman, M. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature Biotechnol. 28, 503–510 (2010)
Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005)
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F. & Mattick, J. S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA 105, 716–721 (2008)
Young, R. S. et al. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol. Evol. 4, 427–442 (2012)
Nam, J. W. & Bartel, D. Long non-coding RNAs in C. elegans. Genome Res. (2012)
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H. & Bartel, D. P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550 (2011)
Chow, J. C., Yen, Z., Ziesche, S. M. & Brown, C. J. Silencing of the mammalian X chromosome. Annu. Rev. Genomics Hum. Genet. 6, 69–92 (2005)
Sleutels, F., Zwart, R. & Barlow, D. P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813 (2002)
Dinger, M. E. et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18, 1433–1445 (2008)
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007)
Ørom, U. A. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58 (2010)
Cesana, M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011)
Chodroff, R. A. et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 11, R72 (2010)
Marques, A. C. & Ponting, C. P. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 10, R124 (2009)
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011)
Kutter, C. et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 8, e1002841 (2012)
Hedges, S. B., Dudley, J. & Kumar, S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972 (2006)
Lin, M. F. et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 17, 1823–1836 (2007)
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005)
The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature 135, 56–65 (2012)
Ward, L. D. & Kellis, M. Evidence of abundant purifying selection in humans for recently-acquired regulatory functions. Science 337, 1675–1678 (2012)
Galtier, N. & Duret, L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 23, 273–277 (2007)
Soumillon, M. et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3, 2179–2190 (2013)
Lindblad-Toh, K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011)
The Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Schmidt, D. et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328, 1036–1040 (2010)
Walker, E., Manias, J. L., Chang, W. Y. & Stanford, W. L. PCL2 modulates gene regulatory networks controlling self-renewal and commitment in embryonic stem cells. Cell Cycle 10, 45–51 (2011)
Chambers, I. & Tomlinson, S. R. The transcriptional foundation of pluripotency. Development 136, 2311–2322 (2009)
Stuart, J. M., Segal, E., Koller, D., Kim, S. K. & A Gene-coexpression network for global discovery of conserved genetic modules. Science 302, 249–255 (2003)
Shkumatava, A., Stark, A., Sive, H. & Bartel, D. P. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 23, 466–481 (2009)
Franceschini, A. et al. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013)
Keniry, A. et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature Cell Biol. 14, 659–665 (2012)
Van Dongen, S. & Abreu-Goodger, C. Using MCL to extract clusters from networks. Methods Mol. Biol. 804, 281–295 (2012)
Grant, J. et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487, 254–258 (2012)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 28, 511–515 (2010)
UniProt. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–D75 (2012)
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012)
Flicek, P. et al. Ensembl 2012. Nucleic Acids Res. 40, D84–D90 (2012)
Yanai, I. et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659 (2005)
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011)
R Development Core Team. R: A language and environment for statistical computinghttp://www.r-project.org (R Foundation for Statistical Computing, 2011)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Rhead, B. et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38, D613–D619 (2010)
Kellis, M., Patterson, N., Birren, B., Berger, B. & Lander, E. S. Methods in comparative genomics: genome correspondence, gene identification and regulatory motif discovery. J. Comput. Biol. 11, 319–355 (2004)
Altschul, S. F., Gish, W., Miller, W., Myers, E. & Lipman, D. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
Vilella, A. J. et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19, 327–335 (2009)
Blanchette, M. et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004)
Kong, A. et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–1103 (2010)
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012)
Carninci, P. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genet. 38, 626–635 (2006)
Kozomara, A. & Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011)
Acknowledgements
We thank L. Froidevaux and D. Cortéz for help with genome sequencing, J. Meunier for help with preliminary miRNA analyses, K. Harshman and the Lausanne Genomics Technology Facility for high-throughput sequencing support, I. Xenarios for computational support, S. Bergmann and Z. Kutalik for advice on co-expression analyses. Human embryonic and fetal material was provided by the Joint MRC/Wellcome Trust (grant 099175/Z/12/Z) Human Developmental Biology Resource (http://www.hdbr.org). The computations were performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the SIB Swiss Institute of Bioinformatics. This research was supported by grants from the European Research Council (Starting Independent Researcher Grant 242597, SexGenTransEvolution) and the Swiss National Science Foundation (grant 31003A_130287) to H.K. A.N. was supported by a FEBS long-term postdoctoral fellowship.
Author information
Authors and Affiliations
Contributions
A.N. conceived and performed all biological analyses and wrote the manuscript, with input from all authors. A.N. and M.W. processed RNA-seq data. M.S. and A.L. generated RNA-seq data. T.D. and F.G. collected platypus samples. U.Z. collected opossum samples. J.C.B. provided mouse placenta samples and contributed to H19X analyses. The project was supervised and originally designed by H.K.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 lncRNA evolutionary age and sequence conservation patterns.
a, Exonic sequence conservation (mean placental PhastCons score), for random intergenic regions, lncRNA maximum evolutionary age classes, coding and untranslated exons of protein-coding genes. b, Mean DAF of autosomal non-CpG SNPs segregating in African populations (1000 Genomes project26). Intergenic SNPs were randomly drawn in regions matching lncRNA recombination rates (Methods). c, Mean DAF for the four classes of mutation orientation (W to S (W→S) or AT to GC; S to W (S→W) or GC to AT; W to W (W→W), or AT to AT; and S to S (S→S), or GC to GC) for autosomal non-CpG SNPs found in primate-specific (age 25 Myr) lncRNA exonic regions (blue) or in intergenic regions with matching recombination rates (grey). The W→S and S→W mutation classes are known to be affected by GC-biased gene conversion. d, Same as c but for lncRNAs that are found close to (left panel, maximum distance 10 kb) or far from (right panel, minimum distance 50 kb) Ensembl-annotated coding or noncoding genes. e, Mean placental PhastCons score for promoter regions (1 kb upstream) of lncRNA minimum evolutionary age classes (beige) and protein-coding genes (blue). f, Mean placental PhastCons score for promoter regions (1 kb upstream) of lncRNA maximum evolutionary age classes (beige) and protein-coding genes (blue). Error bars, 95% confidence intervals based on 100 bootstrap resampling replicates.
Extended Data Figure 2 lncRNA expression patterns in four tetrapod species.
a, Proportions of genes with observed maximum expression in different organs for mouse protein-coding genes, old lncRNAs (shared across at least two species) and young lncRNAs (species-specific). b, Tissue-specificity index, for the same classes of mouse genes. Values close to 1 represent high tissue specificity. c, Distribution of the maximum expression level (log2-transformed RPKM). d–f, Same as a–c but for the opossum. g–i, Same as a–c but for the platypus. j–l, Same as a–c but for the chicken.
Extended Data Figure 3 Transcription-factor binding at lncRNA promoters.
a, Comparison between the frequencies of in silico-predicted transcription-factor (TF)-binding sites in lncRNA promoters (2 kb upstream) and in random intergenic regions. b, Comparison between the frequencies of in silico-predicted TF-binding sites in lncRNA and protein-coding gene promoters (2 kb upstream). Homeobox TFs are shown in blue. c, Comparison between the frequencies of experimentally determined (ChIP-seq ENCODE) TF-binding sites in lncRNA promoters (2 kb upstream) and in random intergenic regions. d, Comparison between the frequencies of experimentally determined (ChIP-seq ENCODE) predicted TF-binding sites in lncRNA and protein-coding gene promoters (2 kb upstream). e, Frequency of binding (Encode ChIP-seq data) for OCT4 (also known as POU5F1). f, g, Proportion of HNF4A- CEBPA-binding events shared between human and mouse, for random intergenic regions, lncRNA (321 lncRNAs with binding events and liver expression, supported by CAGE data) and protein-coding gene promoters (5 kb upstream).
Extended Data Figure 4 Evolution of lncRNA expression patterns.
a, Percentage of human lncRNAs (found in antisense of protein-coding genes) that have transcription evidence in other species, as a function of the divergence time. Transcription evidence was assessed in a pool of brain and testes strand-specific RNA-seq data, for 2,535 human antisense lncRNAs that had 1–1 orthologues in at least one other species and transcription evidence in human (Methods). b, Spearman correlation of human and mouse expression levels, in different tissues. The boxplots represent the variation observed in 100 bootstrap replicates. c, Proportion of human organ-specific protein-coding genes (tissue-specificity index >0.9, RPKM >0.1) for which the organ specificity is shared across primates. Red lines, random expectation of shared organ specificity; horizontal black line, average conserved specificity for all organs. d, Proportion of human organ-specific lncRNAs (minimum evolutionary age >90 Myr, tissue-specificity index >0.9, RPKM >0.1) for which the organ specificity is shared across eutherians. Red lines, random expectation of shared organ specificity; horizontal black line, average conserved specificity for all organs. e, Same as c, conservation across eutherian species. f, Principal component analysis of lncRNA expression levels for families of eutherian 1–1 orthologues. g, Principal component analysis of protein-coding gene expression levels for families of eutherian 1–1 orthologues.
Extended Data Figure 5 Characteristics of the evolutionarily conserved co-expression network.
a, Proportion of activation/inhibition relationships annotated in the String database, for positive and negative co-expression network connections. b, Gene expression levels (maximum over all available sample and species for each co-expression network node) for different network connectivity classes. c, Gene expression levels (maximum over all available sample and species for each co-expression network node) for connected lncRNAs, transcription factors (TFs) and non-TF protein-coding genes. d, Network connectivity (node degree) for lncRNAs (black), transcription factors (medium grey) and for non-transcription factors protein-coding genes (light grey). Top, raw data; bottom, after correcting for expression level differences. e, Difference between observed and expected proportions of connections in cis, for lncRNAs (red), protein-coding genes (blue) and for genes found in HOX clusters (black). The expected proportions were computed through randomizations (Methods).
Extended Data Figure 6 Expression patterns and sequence evolution of H19X-associated miRNAs.
a, Distribution of the average embedded miRNA density (miRNA hairpins per kb, in the gene body or 10 kb downstream), for genes that are positively connected with each network node. Red arrow, average miRNA density for genes that are positively connected with H19. b, Maximum likelihood reconstruction of the phylogeny of the ancient H19X-associated miRNA family (representative members miR-503, miR-322, miR-424, miR-15c, miR-16c). miRNAs associated with H19X are displayed in red (subfamily containing miR-503 and miR-16c) and blue (subfamily containing miR-424, miR-322 and miR-15c). miRNA names are derived from miRBase where available, including three-letter species abbreviations. Hsa, Homo sapiens; Mdo, Monodelphis domestica (opossum); Mml, Macaca mulatta (macaque); Mmu, Mus musculus (mouse); Oan, Ornithorhynchus anatinus (platypus); Gga, Gallus gallus (chicken), Xtr, Xenopus tropicalis. Ensembl identifiers are given for two opossum miRNAs. c, Expression pattern of the mouse miRNA mmu-miR-322, associated with H19X. The expression level was computed as the number of uniquely mapping reads per miRNA, after resampling the same number of reads per tissue. d, Same as c but for the mouse miRNA mmu-miR-351.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion, Supplementary Methods and additional references. (PDF 359 kb)
Supplementary Table 1
This Supplementary Table contains information for the RNA-seq samples used in this study. (XLSX 76 kb)
Supplementary Table 2
Node and edge identifiers for the co-expression network. (XLSX 15528 kb)
Supplementary Table 3
This Supplementary Table contains the list of protein-coding genes, which have an excess of connections in cis in the co-expression network. (XLSX 181 kb)
Supplementary Table 4
MCL clusters determined for the co-expression network and the GO enrichment results for each cluster. (XLSX 263 kb)
Supplementary Tables 5 and 6
This zipped file contains Supplementary Tables 5 and 6. Supplementary Table 5 shows results of the GO enrichment analysis for each lncRNA node in the co-expression network and Supplementary Table 6 contains the list of miRNAs associated with H19X in each species. (ZIP 4765 kb)
Supplementary Data 1
This Supplementary Dataset contains the lncRNA annotations used in this study. (ZIP 20104 kb)
Supplementary Data 2
This Supplementary Dataset contains information for homologous lncRNA families. (ZIP 24012 kb)
Supplementary Data 3
This Supplementary Dataset contains expression level estimates for lncRNAs and for Ensembl-annotated protein-coding genes. (ZIP 25470 kb)
Supplementary Data 4
This Supplementary Dataset contains miRNA expression values for 5 species. (ZIP 21311 kb)
Rights and permissions
About this article
Cite this article
Necsulea, A., Soumillon, M., Warnefors, M. et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014). https://doi.org/10.1038/nature12943
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12943
This article is cited by
-
Genome-scale pan-cancer interrogation of lncRNA dependencies using CasRx
Nature Methods (2024)
-
Deep Conservation and Unexpected Evolutionary History of Neighboring lncRNAs MALAT1 and NEAT1
Journal of Molecular Evolution (2024)
-
Genome-wide analysis of RNA-chromatin interactions in lizards as a mean for functional lncRNA identification
BMC Genomics (2023)
-
‘Lnc-ing’ Treg cells to the aging liver
Nature Aging (2023)
-
lncRNA miR4458HG modulates hepatocellular carcinoma progression by activating m6A-dependent glycolysis and promoting the polarization of tumor-associated macrophages
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.