Abstract
Cells differentiate when transcription factors bind accessible cis-regulatory elements to establish specific gene expression programs. In differentiating embryonic stem cells, chromatin at lineage-restricted genes becomes sequentially accessible1,2,3,4, probably by means of ‘pioneer’ transcription factor activity5, but tissues may use other strategies in vivo. Lateral inhibition is a pervasive process in which one cell forces a different identity on its neighbours6, and it is unclear how chromatin in equipotent progenitors undergoing lateral inhibition quickly enables distinct, transiently reversible cell fates. Here we report the chromatin and transcriptional underpinnings of differentiation in mouse small intestine crypts, where notch signalling mediates lateral inhibition to assign progenitor cells into absorptive or secretory lineages7,8,9. Transcript profiles in isolated LGR5+ intestinal stem cells10 and secretory and absorptive progenitors indicated that each cell population was distinct and the progenitors specified. Nevertheless, secretory and absorptive progenitors showed comparable levels of H3K4me2 and H3K27ac histone marks and DNase I hypersensitivity—signifying accessible, permissive chromatin—at most of the same cis-elements. Enhancers acting uniquely in progenitors were well demarcated in LGR5+ intestinal stem cells, revealing early priming of chromatin for divergent transcriptional programs, and retained active marks well after lineages were specified. On this chromatin background, ATOH1, a secretory-specific transcription factor, controls lateral inhibition through delta-like notch ligand genes and also drives the expression of numerous secretory lineage genes. Depletion of ATOH1 from specified secretory cells converted them into functional enterocytes, indicating prolonged responsiveness of marked enhancers to the presence or absence of a key transcription factor. Thus, lateral inhibition and intestinal crypt lineage plasticity involve interaction of a lineage-restricted transcription factor with broadly permissive chromatin established in multipotent stem cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006)
Wamstad, J. A. et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151, 206–220 (2012)
Rada-Iglesias, A. et al. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11, 633–648 (2012)
Xie, W. et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148 (2013)
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011)
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999)
van Es, J. H. et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005)
Pellegrinet, L. et al. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240 (2011)
Stamataki, D. et al. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS ONE 6, e24484 (2011)
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007)
Yang, Q., Bermingham, N. A., Finegold, M. J. & Zoghbi, H. Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294, 2155–2158 (2001)
Kim, T. H. & Shivdasani, R. A. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J. Biol. Chem. 286, 11427–11433 (2011)
VanDussen, K. L. & Samuelson, L. C. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev. Biol. 346, 215–223 (2010)
Shroyer, N. F. et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488 (2007)
Milano, J. et al. Modulation of notch processing by γ-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82, 341–358 (2004)
Kazanjian, A., Noah, T., Brown, D., Burkart, J. & Shroyer, N. F. Atonal homolog 1 is required for growth and differentiation effects of notch/γ-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology 139, 918–928 (2010)
van Es, J. H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature Cell Biol. 14, 1099–1104 (2012)
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007)
Verzi, M. P. et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19, 713–726 (2010)
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007)
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012)
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011)
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010)
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008)
Klisch, T. J. et al. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc. Natl Acad. Sci. USA 108, 3288–3293 (2011)
Ferrell, J. E., Jr Bistability, bifurcations, and Waddington’s epigenetic landscape. Curr. Biol. 22, R458–R466 (2012)
Schwitalla, S. et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38 (2013)
Buczacki, S. J. et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69 (2013)
Rose, M. F., Ahmad, K. A., Thaller, C. & Zoghbi, H. Y. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc. Natl Acad. Sci. USA 106, 22462–22467 (2009)
Kaaij, L. T. et al. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 14, R50 (2013)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zang, C. et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 (2009)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nature Protocols 7, 1728–1740 (2012)
Liu, T. et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83 (2011)
Irizarry, R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003)
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, http://dx.doi.org/10.2202/1544-6115.1027 (2004)
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002)
El Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004)
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001)
ENCODE Project Consortium A user’s guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9, e1001046 (2011)
Acknowledgements
This work was supported by NIH grants R01DK082889 and R01DK081113 (R.A.S.), K99DK095983 (T.-H.K.), K01DK088868 (M.V.), and P50CA127003; a North American Neuroendocrine Tumor Society fellowship (T.-H.K.); and the Caring For Carcinoid Foundation (R.A.S.). We thank S. Robine for villin-CreER-T2 mice; T. Honjo and S. Artavanis-Tsakonas for Rbpjfl mice; J. Johnson for ATOH1 antibody; D. Podolsky for TFF3 antibody; and M. Brown and P. Cejas for discussions.
Author information
Authors and Affiliations
Contributions
T.-H.K. conceived and designed experiments, collected and analysed data, and drafted the manuscript; F.L. performed computational analyses; I.F.-N. performed some mouse experiments; L.-L.H, A.L., K.N. and M.V. prepared some ChIP-seq libraries; H.L. supervised computational analyses; R.A.S. conceived and supervised the study, analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Isolation of intestinal stem cells and other intestinal cells.
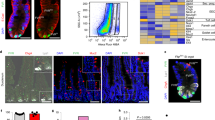
a, Crypt-villus unit structure and components of crypt lateral inhibition. Terminally differentiated enterocytes and secretory cells line intestinal villi and depend on self-renewing crypt populations of LGR5+ intestinal stem cells, bipotential (Bi-Pro), and specified secretory (Sec-Pro) and enterocyte (Ent-Pro) progenitors. Stochastic ATOH1 expression in one Bi-Pro induces secretory differentiation and delta ligand expression, allowing notch signal delivery to neighbouring cells, which use HES1 to repress Atoh1 and become enterocytes. b, Intestinal epithelial cell isolation strategies. LGR5+ intestinal stem cells were isolated from Lgr5GFP-creER mice10; Sec-Pro and goblet cells were isolated from Rbpj−/− intestines7; additional Sec-Pro cells were collected from DBZ-treated wild-type mouse crypts; Ent-Pro and enterocytes were collected from Atoh1−/− intestines14. c, Crypt cells expressing high LGR5/GFP were isolated by flow cytometry and pooled from 12 mice. Gates to distinguish GFPhi intestinal stem cells from their GFPlo and GFP− progeny are shown in the representative scatter plot. d, e, Alcian blue staining of Atoh1−/− intestines at low (d) and high (e) magnification, demonstrating global absence of goblet cells. Inset in e shows the boxed region at higher magnification. f, Rbpjfl/fl;villin-creER intestines rendered null for notch activity 6 days after tamoxifen injection, showing severe goblet cell metaplasia, comparable to DBZ-treated wild-type intestines (See Extended Data Fig. 2d). Scale bars, 50 µm. Duplicate samples were examined from three (Rbpj−/−) or seven (Atoh1−/−) mice.
Extended Data Figure 2 Enrichment of Sec-Pro by chemical inhibition of notch signalling.
a, Quantitative RT–PCR data showing peak Atoh1 expression ∼38 h after treatment of wild-type mice with dibenzazepine (DBZ). b–d, Low magnification views show no increase in mature, Alcian-blue-avid goblet cells 38 h after DBZ treatment (b), appearance of goblet cells in villus bases and crypts 72 h after DBZ treatment (c), and migration of these cells to occupy villi almost fully within 6 days (d). Inset in b shows a magnified view of the boxed area. e, Within 72 h of DBZ treatment, crypt cells virtually cease to replicate (E), retain ATOH1 (C), and stain with Alcian blue (A), a sign of goblet cell maturation. By 120 h after DBZ exposure, these ATOH1+ post-mitotic goblet cells occupy short villi (B, D, F). Scale bars, 50 μm. Tissue analyses were done in duplicate on seven (DBZ+38 h) or three (DBZ+72 h, DBZ+144 h) mice.
Extended Data Figure 3 Distinct expression profiles in LGR5+ intestinal stem cells, Sec-Pro, Ent-Pro, and villus enterocytes.
Differential transcripts in purified intestinal cell types, arranged by k-means clustering of normalized expression values from Affymetrix microarrays. Representative known genes in each cell type are listed alongside the Gene Ontology functions most enriched in each group (false discovery rates in parentheses), determined using Gene Set Enrichment Analysis tools (http://www.broadinstitute.org/gsea/msigdb/annotate.jsp).
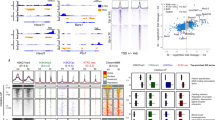
Extended Data Figure 4 Summary of ChIP-seq and DHS-seq experiments.
a, Summary of sites identified by MACS II in ChIP-seq and DHS-seq data from intestinal cell populations. b, Distribution of sites marked in promoters (≤2 kb upstream or ≤1 kb downstream of any TSS), coding regions, introns and intergenic regions. c, Correlation analysis with Spearman’s rank coefficient (left) and sample traces (right) of H3K4me2-marked enhancers in independent ChIP-seq replicates of Ent-Pro, showing very high concordance. d, H3K4me2 ChIP-seq profiles at arbitrarily selected regions of mouse chromosome 17 illustrate the highly concordant signals in Sec-Pro produced by Rbpj loss (top traces) or DBZ treatment (bottom traces), as heat maps and clustering analysis (Fig. 2a, b) demonstrate throughout the genome.
Extended Data Figure 5 Accessible, marked enhancers in intestinal crypt progenitors.
a, b, ChIP-seq and DHS-seq traces at sample secretory loci illustrate selective intron marking in goblet cells at Tpsg1 (a), representing the profiles in group 2 enhancers (Fig. 2a), and highly similar distribution of H3K4me2, H3K27ac and DHS signals (within dotted, asterisked lines) across cell populations at Foxa1 (b), representing the prevalent group 1 pattern. c, Heat maps of DHS at 116,324 to 139,312 enhancers showing accessible chromatin in Sec-Pro and Ent-Pro cells, compared to DNase I insensitivity at the same sites in adipocytes, cardiac and skeletal muscle (data from the ENCODE Project Consortium41), and lack of chromatin access in crypt progenitors at enhancers that are active in these heterologous cells. d, At a few secretory loci, such as Spdef, signs of enhancer activity and access are higher in Rbpj−/− Sec-Pro or goblet cells than in other intestinal cells. Asterisked intervals mark ATOH1 binding regions.
Extended Data Figure 6 ATOH1 transcriptional control of secretory differentiation.
a, Model for cell-specific ATOH1 binding to permissive chromatin in crypt progenitors. b, Phastcons plot showing high evolutionary conservation of ATOH1-occupied sites in Sec-Pro cells and maximal enrichment of the consensus ATOH1 recognition motif near their peaks. c, Composite profiles of average H3K4me density near ATOH1 binding sites in Sec-Pro cells (left) and mouse brain (right). H3K4me marks are highly correlated with ATOH1 occupancy in the same, but not in the heterologous, tissue, as representative ChIP-seq data illustrate below. d, e, Examples of ATOH1 occupancy at loci for key secretory lineage transcription factors and secretory-specific genes: Nkx2-2 and Foxa2 (enteroendocrine cells), Xbp1 (Paneth cells), Tff3 (goblet cells), Spink4 and Hgfac (goblet and Paneth cells) and Sstr1 (endocrine cells). ATOH1 does not bind near Ent-Pro-specific genes, as shown at a sample locus, Bcl2l15. f, Schematic representation of ATOH1’s dual function in direct control of two gene groups: delta ligands for lateral inhibition and key secretory-specific transcription factors to promote secretory lineage differentiation. g, Relation of ATOH1 occupancy at enhancers with Sec-Pro-specific transcription. Bins of 200 genes, ranked by differential expression in Sec-Pro (left) or Ent-Pro (right), are represented along the x axis, and normalized ATOH1 binding in Sec-Pro (red) or brain (blue) at enhancers within 20 kb of the genes in each bin is represented on the y axis. The graphs show the first (Sec-Pro-specific), middle (no differential expression), and last (Ent-Pro-specific) three bins of 200 genes each. Error bars indicate standard errors of the mean and pair-wise t-tests support a role for ATOH1 in activating Sec-Pro genes, without an overt role in silencing Ent-Pro genes.
Extended Data Figure 7 ATOH1 binding at H3K4me2-marked enhancers.
Representation of ATOH1 binding sites in relation to the heat map of H3K4me2 marking at 57,481 non-promoter sites (from Fig. 2a), showing that ATOH1 occupies an abundance of sites marked in all crypt populations. Only in mature villus cells does ATOH1 preferentially bind enhancers that are selectively marked in goblet cells.
Extended Data Figure 8 ATOH1-dependent lineage conversion.
a–d, BrdU tracing 1 and 3 days after label injection, showing similar turnover of intestinal epithelial cells in wild-type and Atoh1-null intestines in the absence of Notch inhibition. Nearly all the label is in the villus base at 1 day and in the top halves of villi at 3 days. e, Lineage tracing 3 days after induction of Cre activity by tamoxifen in Lgr5cre-ER;Rosa26YFP mice (left) or by RU486 in Atoh1cre-PR;Rosa26YFP mice (right). YFP is detected in numerous whole crypts (except Paneth cells) in the former, and never in crypts, ribbons or cell pairs in the latter, confirming absence of leaky CrePR expression in multipotent progenitors. f, YFP signal in Atoh1cre-PR;Rosa26YFP mice 3 days after RU486-induced Cre activation is confined to scattered ChgA+ enteroendocrine and Muc2+ goblet cells. g, Two additional representative examples of YFP-traced cell groups that had converted into TFF3-negative enterocytes after Atoh1 deletion in secretory cells. Arrows indicate rare (∼5%, see text) YFP+ cells that still express TFF3. Scale bars, 50 μm. Tissues were analysed in duplicate from four (WT, Atoh1−/−, Atohcre-PR/+;Rosa26YFP) or six (Lgr5cre-ER;Rosa26YFP) mice.
Supplementary information
Supplementary Table 1
This file shows transcripts enriched in Sec-Pro and Ent-Pro. (XLSX 58 kb)
Supplementary Table 2
This file shows genes and TF motifs associated with cell type-specific enhancers. (XLSX 567 kb)
Rights and permissions
About this article
Cite this article
Kim, TH., Li, F., Ferreiro-Neira, I. et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 506, 511–515 (2014). https://doi.org/10.1038/nature12903
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12903
This article is cited by
-
Cell-fate conversion of intestinal cells in adult Drosophila midgut by depleting a single transcription factor
Nature Communications (2024)
-
Epigenetic control of cellular crosstalk defines gastrointestinal organ fate and function
Nature Communications (2023)
-
IFNγ-Stat1 axis drives aging-associated loss of intestinal tissue homeostasis and regeneration
Nature Communications (2023)
-
Advanced Progression for the Heterogeneity and Homeostasis of Intestinal Stem Cells
Stem Cell Reviews and Reports (2023)
-
The cellular niche for intestinal stem cells: a team effort
Cell Regeneration (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.