Abstract
CCAAT/enhancer binding protein-α (C/EBPα) induces transdifferentiation of B cells into macrophages at high efficiencies and enhances reprogramming into induced pluripotent stem (iPS) cells when co-expressed with the transcription factors Oct4 (Pou5f1), Sox2, Klf4 and Myc (hereafter called OSKM)1,2. However, how C/EBPα accomplishes these effects is unclear. Here we find that in mouse primary B cells transient C/EBPα expression followed by OSKM activation induces a 100-fold increase in iPS cell reprogramming efficiency, involving 95% of the population. During this conversion, pluripotency and epithelial–mesenchymal transition genes become markedly upregulated, and 60% of the cells express Oct4 within 2 days. C/EBPα acts as a ‘path-breaker’ as it transiently makes the chromatin of pluripotency genes more accessible to DNase I. C/EBPα also induces the expression of the dioxygenase Tet2 and promotes its translocation to the nucleus where it binds to regulatory regions of pluripotency genes that become demethylated after OSKM induction. In line with these findings, overexpression of Tet2 enhances OSKM-induced B-cell reprogramming. Because the enzyme is also required for efficient C/EBPα-induced immune cell conversion3, our data indicate that Tet2 provides a mechanistic link between iPS cell reprogramming and B-cell transdifferentiation. The rapid iPS reprogramming approach described here should help to fully elucidate the process and has potential clinical applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hanna, J. et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133, 250–264 (2008)
Xie, H., Ye, M., Feng, R. & Graf, T. Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676 (2004)
Kallin, E. M. et al. Tet2 facilitates the derepression of myeloid target genes during CEBPα-induced transdifferentiation of pre-B cells. Mol. Cell 48, 266–276 (2012)
Buganim, Y. et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209–1222 (2012)
Hanna, J. et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 (2009)
Orkin, S. H. & Hochedlinger, K. Chromatin connections to pluripotency and cellular reprogramming. Cell 145, 835–850 (2011)
Costa, Y. et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 (2013)
Doege, C. A. et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–655 (2012)
Rais, Y. et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502, 65–70 (2013)
Di Tullio, A. et al. CCAAT/enhancer binding protein α (C/EBPα)-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc. Natl Acad. Sci. USA 108, 17016–17021 (2011)
Carey, B. W., Markoulaki, S., Beard, C., Hanna, J. & Jaenisch, R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nature Methods 7, 56–59 (2010)
Eminli, S. et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nature Genet. 41, 968–976 (2009)
Porse, B. T. et al. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo . Cell 107, 247–258 (2001)
Golipour, A. et al. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11, 769–782 (2012)
Li, R. et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 (2010)
Samavarchi-Tehrani, P. et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 (2010)
Liu, X. et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nature Cell Biol. 15, 829–838 (2013)
Gao, Y. et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 12, 453–469 (2013)
Ang, Y. S. et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197 (2011)
Kulessa, H., Frampton, J. & Graf, T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9, 1250–1262 (1995)
Laiosa, C. V., Stadtfeld, M., Xie, H., de Andres-Aguayo, L. & Graf, T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25, 731–744 (2006)
Feng, R. et al. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc. Natl Acad. Sci. USA 105, 6057–6062 (2008)
Zhang, D. E. et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl Acad. Sci. USA 94, 569–574 (1997)
Begay, V., Smink, J. & Leutz, A. Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol. Cell. Biol. 24, 9744–9751 (2004)
Guilbert, L., Robertson, S. A. & Wegmann, T. G. The trophoblast as an integral component of a macrophage-cytokine network. Immunol. Cell Biol. 71, 49–57 (1993)
Cheng, J. et al. An extensive network of TET2-targeting microRNAs regulates malignant hematopoiesis. Cell Rep. 5, 471–481 (2013)
Di Stefano, B. et al. A microRNA-based system for selecting and maintaining the pluripotent state in human induced pluripotent stem cells. Stem Cells 29, 1684–1695 (2011)
Di Tullio, A. & Graf, T. C/EBPα bypasses cell cycle-dependency during immune cell transdifferentiation. Cell Cycle 11, 2739–2746 (2012)
van Oevelen, C. et al. A role for mammalian Sin3 in permanent gene silencing. Mol. Cell 32, 359–370 (2008)
Tilgner, H. et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 22, 1616–1625 (2012)
Acknowledgements
We thank L. Batlle for generating the chimaeric mice, M. Stadtfeld, D. Egli, P. Cosma, G. Filion, B. Lehner and J. Valcarcel for critical reading of the manuscript, V. Broccoli, P. Muñoz and S. Aznar Benitah for constructs and antibodies and H. Schoeler for the Oct4 reporter mice. This work was supported by the Ministerio de Educacion y Ciencia, SAF.2007-63058 and AGAUR 2009 SGR768. B.D.S. holds a La Caixa International PhD Fellowship.
Author information
Authors and Affiliations
Contributions
T.G. and B.D.S. conceived the idea for this project, designed the experiments and wrote the paper. B.D.S. performed the cell culture, animal experiments and molecular biology and B.D.S. and J.L.S. the methylation analyses. J.L.S. performed the studies on Tet2 and J.L.S and C.v.O. the ChIP experiments. S.C., E.M.K. and D.T. were responsible for the bioinformatics and G.P.V. and M.B. for the DNase I experiments and analyses. J.L. provided the Tet2 overexpressing plasmids.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Single cell reprogramming.
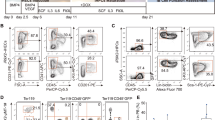
a, Bα′ cells were plated as single cells in 96-well plates on OP9 feeders in medium conditioned with IL-7, IL-15 and IL-4 cytokines. b, Representative Nanog-positive (upper panels) and Nanog-negative (lower panels) iPS cell clones in 96-well plates (scale bar, 100 μm). c, Efficiency of reprogramming obtained from two independent experiments.
Extended Data Figure 2 C/EBPα mutant and cell viability during iPS cell reprogramming.
a, E2 has no effect on reprogramming efficiency and ectopic Oct4 expression. Error bars indicate s.d. (n = 3). b, Effect of a C/EBPα mutant for the DNA binding domain on iPS cell reprogramming efficiency. Student’s t-test ***P < 0.001 relative to control. Error bars indicate s.d. (n = 3). c, Histogram showing percentages of live cells during the first 3 days of iPS cell reprogramming as analysed by FACS. Dead cells were scored as positive for DAPI staining. d, Effect of 18 h C/EBPα pulses after OSKM induction. Error bars indicate s.d. (n = 3).
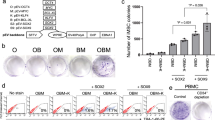
Extended Data Figure 3 Properties of αiPS cells.
a, Heat maps showing gene expression profiles of B cells, ES cells, αiPS cells (clone 22) and iPS cells (clone 26) derived from Bα′ and B cells, respectively. Genes were selected based on a >4-fold difference between B cells and ES cells. b, Micrographs (×10) of αiPS cell clone 22 growing on MEF feeders, showing colonies of ES-like morphology (bright field) and expression of Nanog (green) and Oct4 (red). c, Embryoid bodies generated from αiPS cell clone 22 (bright field) differentiated into Tuj1-positive cells (ectoderm), Foxa2-positive cells (endoderm) and SMA expressing cells (mesoderm). Original magnification, ×10. d, Section of a teratoma obtained from αiPS clone 22 stained with haematoxylin and eosin, showing differentiation into the major germ layers Original magnification, ×10. e, Proportion of chimaeric mice obtained after injection of CD1 blastocysts with three different αiPS cell clones.
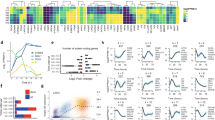
Extended Data Figure 4 Pluripotency gene regulation during iPS cell reprogramming.
a, Unsupervised hierarchical clustering analysis of gene expression array data. b, Agilent gene expression kinetics (log2 expression units) of pluripotency genes from Bα′+OSKM cells (red shaded panels), αiPS cells and ES cells (white panels) and B+OSKM cells (green shaded panels). c, qRT–PCR confirming array results for Dnmt3b and Zfp42 (Rex1) genes. Normalized against Pgk expression. Error bars indicate s.d. (n = 3). d, Agilent gene expression kinetics of late transition genes. e, Oct4–GFP expression during iPS cell reprogramming.
Extended Data Figure 5 Changes in the expression of mesenchymal–epithelial transition genes.
a, Agilent gene expression kinetics of mesenchymal–epithelial transition (MET) genes. b, E-cadherin expression by FACS at different times after induction. Isotype controls are shown in black; antibody-stained cells in red and green. c, GO analysis for genes upregulated >2-fold in Bα′ cells compared with B cells. d, GO analysis for genes downregulated >2-fold in Bα′ cells compared with B cells. e, GO analysis of genes upregulated >2-fold in Bα′ cells compared with B cells and subsequently downregulated >2-fold at 48 h.
Extended Data Figure 6 Changes in the expression of B cell and macrophage genes.
a, Heat map of expression Affymetrix arrays with 263 macrophage and 83 B-cell-specific genes during C/EBPα-induced transdifferentiation of B cells and heat maps of the same genes using data from the Agilent expression array in Fig. 2. b, Agilent gene expression kinetics of selected macrophage and B-cell transcription factors. c, RNA-seq data for two representative macrophage and B-cell-specific genes each. d, FACS profiles of E-cadherin (Cdh1) expression, combined with either CSF-1R (upper panels) or Mac1 (lower panels) staining. e, Reprogramming potential of E-cadherin+/Mac1− and E-cadherin+/Mac1+ cells. Error bars indicate s.d. (n = 3).
Extended Data Figure 7 ChIP-seq and RNA-seq analysis.
a, Number of upregulated and downregulated genes (>1.5-fold change) after a pulse of C/EBPα. b, Result of gene set enrichment analysis on RNA-seq data (ranked by fold change) for selected significantly enriched gene sets (from MSigDB cp.v4 database). c, Circular visualization of genomic distribution of differentially expressed genes (RNA-seq) and C/EBPα binding sites (ChIP-seq). d, Distribution of C/EBPα peaks into genomic features, on the basis of Refseq mm9 annotation. e, Fraction of upregulated (red chart) and downregulated genes (blue) after a pulse of C/EBPα showing a peak at 50 kb from TSS. f, Variation of expression after C/EBPα pulse of nearest genes to peaks. g, Selected gene sets significantly enriched in genes adjacent to C/EBPα peaks based on hypergeometric tests (from MSigDB cp.v4 database). Bonferoni procedure, P value <0.05.
Extended Data Figure 8 Tet2 expression in B cells and MEFs.
a, RT–qPCR of Tet2, pluripotency and MET genes after induction of OSKM in MEFs pulsed or not with C/EBPα. Error bars indicate s.d. (n = 3). b, c, hMeDIP and Tet2 binding at regulatory regions of the Tet2 target gene Hal1 (ref. 3) and Sall4. d, Tet2 localization in B cells pulsed with C/EBPα for 72 h. e, Oestradiol treatment has no effect on cellular localization of Tet2 protein in B cells (scale bar, 30 μm). f, Cellular localization of Tet2 protein in MEFs treated or not with E2 (scale bar, 100 μm).
Extended Data Figure 9 DNase I sensitivity and gene expression summary.
a, DNase I sensitivity at regulatory regions in B cells and Bα′ cells (data from Fig. 4a) and B cells pulsed for 72 h determined by enzyme titration. Error bars indicate s.d. (n = 3). Student’s t-test *P < 0.05 relative to control. b, CpGs analysed for methylation by bisulphite sequencing in the Pou5f1 and Nanog promoters. Filled rectangles represent methylated CpGs, empty rectangles unmethylated residues. c, Summary of relevant gene expression changes during the transition from B cells to Bα′ cells and Bα′ cells to αiPS cells.
Extended Data Figure 10 Transcription factor and cell type specificity.
a, b, B+OSKM cells were infected with a retrovirus expressing GATA1 or left uninfected, induced with doxycycline and scored for 12 d.p.i. alkaline phosphatase+ and Nanog+ colonies. Error bars indicate s.d. (n = 3). c, qRT–PCR for megakaryocyte/erythroid-restricted genes in B cells infected with GATA1 retrovirus for 3 days. Error bars indicate s.d. (n = 3). d, e, Alkaline phosphatase+ colonies at 15 d.p.i. in OSKM MEFs infected with a retrovirus expressing Mash1 or mock infected, and Nanog+ colonies counted. Error bars indicate s.d. (n = 3). f, g, Effect of MyoD expression on iPS cell reprogramming of MEFs. Cells were infected with MyoD-ER and sequentially induced with E2 to activate MyoD and doxycycline to activate OSKM. Representative plates with alkaline phosphatase+ iPS cell colonies 15 d.p.i. of MEFs pre-induced for 24, 48 or 72 h or continuously with E2. Error bars indicate s.d. (n = 3). h, Expression of muscle-restricted genes in OSKM MEFs 3 days after MyoD-ER induction. Error bars indicate s.d. (n = 3). i, Effects of C/EBPα and C/EBPβ in B+OSKM cells. Student’s t-test ***P < 0.001 relative to control. Error bars indicate s.d. (n = 3). j, Effect of an 18-h C/EBPα pulse in pre-T+OSKM cells on the formation of 12 d.p.i. Nanog+ colonies. Student’s t-test *P < 0.05 relative to control. Error bars indicate s.d. (n = 3). k, Effects of C/EBPα, PU.1 and their combination in MEFs. Student’s t-test *P < 0.05 relative to control. Error bars indicate s.d. (n = 3). l, Effect of Pax5 knock down in B-cell reprogramming. Student’s t-test *P < 0.05 relative to control. Error bars indicate s.d. (n = 3).
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-2. (PDF 183 kb)
Rights and permissions
About this article
Cite this article
Di Stefano, B., Sardina, J., van Oevelen, C. et al. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 506, 235–239 (2014). https://doi.org/10.1038/nature12885
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12885
This article is cited by
-
Unraveling the 2,3-diketo-l-gulonic acid-dependent and -independent impacts of l-ascorbic acid on somatic cell reprogramming
Cell & Bioscience (2023)
-
Dynamics of alternative splicing during somatic cell reprogramming reveals functions for RNA-binding proteins CPSF3, hnRNP UL1, and TIA1
Genome Biology (2021)
-
AKT signaling is associated with epigenetic reprogramming via the upregulation of TET and its cofactor, alpha-ketoglutarate during iPSC generation
Stem Cell Research & Therapy (2021)
-
Key features of the POU transcription factor Oct4 from an evolutionary perspective
Cellular and Molecular Life Sciences (2021)
-
Pro-neuronal activity of Myod1 due to promiscuous binding to neuronal genes
Nature Cell Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.