Abstract
Cells of the osteoblast lineage affect the homing1,2 and the number of long-term repopulating haematopoietic stem cells3,4, haematopoietic stem cell mobilization and lineage determination and B cell lymphopoiesis5,6,7. Osteoblasts were recently implicated in pre-leukaemic conditions in mice8,9. However, a single genetic change in osteoblasts that can induce leukaemogenesis has not been shown. Here we show that an activating mutation of β-catenin in mouse osteoblasts alters the differentiation potential of myeloid and lymphoid progenitors leading to development of acute myeloid leukaemia with common chromosomal aberrations and cell autonomous progression. Activated β-catenin stimulates expression of the Notch ligand jagged 1 in osteoblasts. Subsequent activation of Notch signalling in haematopoietic stem cell progenitors induces the malignant changes. Genetic or pharmacological inhibition of Notch signalling ameliorates acute myeloid leukaemia and demonstrates the pathogenic role of the Notch pathway. In 38% of patients with myelodysplastic syndromes or acute myeloid leukaemia, increased β-catenin signalling and nuclear accumulation was identified in osteoblasts and these patients showed increased Notch signalling in haematopoietic cells. These findings demonstrate that genetic alterations in osteoblasts can induce acute myeloid leukaemia, identify molecular signals leading to this transformation and suggest a potential novel pharmacotherapeutic approach to acute myeloid leukaemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Heissig, B. et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of Kit-ligand. Cell 109, 625–637 (2002)
Shiozawa, Y. et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J. Clin. Invest. 121, 1298–1312 (2011)
Calvi, L. M. et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425, 841–846 (2003)
Zhang, J. et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836–841 (2003)
Wu, J. Y. et al. Osteoblastic regulation of B lymphopoiesis is mediated by Gsα-dependent signaling pathways. Proc. Natl Acad. Sci. USA 105, 16976–16981 (2008)
Zhu, J. et al. Osteoblasts support B-lymphocyte commitment and differentiation from hematopoietic stem cells. Blood 109, 3706–3712 (2007)
Chan, C. K. et al. Endochondral ossification is required for haematopoietic stem-cell niche formation. Nature 457, 490–494 (2009)
Walkley, C. R. et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell 129, 1097–1110 (2007)
Raaijmakers, M. H. et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 464, 852–857 (2010)
Glass, D. A. et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev. Cell 8, 751–764 (2005)
Kogan, S. C. et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 100, 238–245 (2002)
Sternberg, A. et al. Evidence for reduced B-cell progenitors in early (low-risk) myelodysplastic syndrome. Blood 106, 2982–2991 (2005)
van de Loosdrecht, A. A. et al. Identification of distinct prognostic subgroups in low- and intermediate-1-risk myelodysplastic syndromes by flow cytometry. Blood 111, 1067–1077 (2008)
Raza, A. & Galili, N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nature Rev. Cancer 12, 849–859 (2012)
The Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013)
Estrach, S., Ambler, C. A., Lo, C. C., Hozumi, K. & Watt, F. M. Jagged 1 is a β-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development 133, 4427–4438 (2006)
Real, P. J. et al. γ-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nature Med. 15, 50–58 (2009)
Baia, G. S. et al. Notch activation is associated with tetraploidy and enhanced chromosomal instability in meningiomas. Neoplasia 10, 604–612 (2008)
Schepers, K. et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell 13, 285–299 (2013)
Delaney, C. et al. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nature Med. 16, 232–236 (2010)
Mercher, T. et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J. Clin. Invest. 119, 852–864 (2009)
Lobry, C. et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J. Exp. Med. 210, 301–319 (2013)
Klinakis, A. et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 473, 230–233 (2011)
Kannan, S. et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J. Exp. Med. 210, 321–337 (2013)
Krebs, L. T. et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343–1352 (2000)
Kumano, K. et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity 18, 699–711 (2003)
Mancini, S. J. et al. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood 105, 2340–2342 (2005)
Lobry, C., Oh, P. & Aifantis, I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J. Exp. Med. 208, 1931–1935 (2011)
Grabher, C., von Boehmer, H. & Look, A. T. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nature Rev. Cancer 6, 347–359 (2006)
Dacquin, R., Starbuck, M., Schinke, T. & Karsenty, G. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev. Dyn. 224, 245–251 (2002)
Harada, N. et al. Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 18, 5931–5942 (1999)
Rached, M. T. et al. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J. Clin. Invest. 120, 357–368 (2010)
Logan, C. Y. & Nusse, R. The Wnt signalling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 (2004)
Rossert, J., Eberspaecher, H. & de Crombrugghe, B. Separate cis-acting DNA elements of the mouse pro-α1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol. 129, 1421–1432 (1995)
Kiernan, A. E., Xu, J. & Gridley, T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2, e4 (2006)
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010)
Tiacci, E. et al. BRAF mutations in hairy-cell leukemia. N. Engl. J. Med. 364, 2305–2315 (2011)
Trifonov, V., Pasqualucci, L., Tiacci, E., Falini, B. & Rabadan, R. SAVI: a statistical algorithm for variant frequency identification. BMC. Systems Biol. http://dx.doi.org/10.1186/1752-0509-7-S2-S2 (2013)
Hubbell, E., Liu, W. M. & Mei, R. Robust estimators for expression analysis. Bioinformatics 18, 1585–1592 (2002)
Barrett, T. et al. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 33, D562–D566 (2005)
Smyth, G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet Mol. Biol. 3, Article 3 (2004)
Simon, R. M. et al. in Design and Analysis of DNA Microarray Investigations (eds Simon, R. M. et al.) 65–94 (Springer, 2003)
van Es, J. H. et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005)
Eghbali-Fatourechi, G. Z. et al. Circulating osteoblast-lineage cells in humans. N. Engl. J. Med. 352, 1959–1966 (2005)
Rubin, M. R. et al. Parathyroid hormone stimulates circulating osteogenic cells in hypoparathyroidism. J. Clin. Endocrinol. Metab. 96, 176–186 (2011)
Manavalan, J. S. et al. Circulating osteogenic precursor cells in type 2 diabetes mellitus. J. Clin. Endocrinol. Metab. 97, 3240–3250 (2012)
Ghosh-Choudhury, N., Harris, M. A., Feng, J. Q., Mundy, G. R. & Harris, S. E. Expression of the BMP 2 gene during bone cell differentiation. Crit. Rev. Eukaryot. Gene Expr. 4, 345–355 (1994)
Rached, M. T. et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 11, 147–160 (2010)
Parfitt, A. M. et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2, 595–610 (1987)
Parfitt, A. M. in Bone Histomorphometry: Techniques and Interpretation (ed. Recker, R. R. ) 143–223 (CRC Press, 1983)
Recker, R. R. et al. Issues in modern bone histomorphometry. Bone 49, 955–964 (2011)
Acknowledgements
The authors are grateful to M. Taketo for providing the Catnb+/lox(ex3) mice; R. Dalla-Favera and G. Karsenty for helpful discussions and critical reading of the manuscript; M. Van Den Brink for providing funding support; D. McMahon and C. A. Zhang for help with statistical analysis; and J. Sha, F. N. Emmons, I. Linkov and J. Pichard for technical assistance. The histology and metabolic unit facility of the Diabetes and Endocrinology Research Center (DERC, NIDDK DK063608-07), the Molecular Pathology facility of the Herbert Irving Cancer Center of Columbia University Medical Center provided help with histological analysis and W. Fantl and G. Nolan for provided flow cytometry conditions for the nuclear β-catenin antibody. This work was supported by the National Institutes of Health (R01 AR054447, and P01 AG032959 and R01 AR055931 to S.K.) and by the Division of Hematologic Oncology, Memorial Sloan-Kettering Cancer Center, New York (to E.B.) and by a European Union Marie Curie Fellowship INTERBONE−FP7−PEOPLE−2011−IRSES−295181 (to A.B.).
Author information
Authors and Affiliations
Contributions
A.K. and S.K. initiated the study and designed the experiments. A.K., J.S.M. and S.K. analysed the data. A.K. carried out most of the experimental work with the help of J.S.M., I.M. and N.L. J.S.M. performed the flow cytometry analysis. H.K., A.L. and R.R. performed whole-exome sequencing analysis. I.M. confirmed exome mutations. N.L. performed immunofluorescence. C.V.R. reviewed and discussed haematopoiesis data and bone marrow transplantations. G.B., D.P. and J.T.-F. performed histology in mouse samples. J.T.-F. and D.P. performed histology in human samples. A.R., S.M., N.G., J.T.-F. and E.B. provided human AML and MDS samples and reviewed and discussed human bone marrow and bone biopsy data. M.V. performed G-banding karyotype analysis. R.F. analysed microarray data. A.B. assisted with mouse flow cytometry experiments. A.K. and S.K. wrote the manuscript. S.K. directed the research. All authors discussed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Anaemia, peripheral blood leukocytosis and monocytosis and deregulated haematopoiesis specific activation of β-catenin in osteoblasts of Ctnnb1CAosb mice.
a, Haematopoietic parameters. White blood cells (WBC), red blood cells (RBC), haemoglobin (HB), haematocrit (HCT), platelets, lymphocytes (LY), neutrophils (NE) and monocytes (MO) in 2-week-old mice. b–k, Bone marrow samples. b, Erythroid cell numbers. c, Representative flow cytometry image showing monocytic/granulocytic (CD11b/Gr1+) subset. d, Numbers of the CD11b+/Gr1+ subset. e, Distribution of LSK (Lineage−Sca+C-kit+) population. f, LSK numbers. g, h, Frequency (g) and percentage (h) of LT-HSCs and ST-HSCs. i, Numbers of LSK+/ FLT3+ cells. j, Myeloid progenitor profile by CD34 versus FcgRII/III analysis of electronically gated Lin−Sca-1−c-Kit+ bone marrow cells. k, Numbers of myeloid progenitor populations. l, Spleen weight. m, Extramedullar haematopoiesis in the liver of 3-weeks-old Ctnnb1CAosb mice indicated by megakaryocytes (black arrow), myeloid (white arrow) and rare erythroid precursors (dotted arrow). n–p Percentage of Ter119+ (n), CD11b+/Gr1+ (o) and myeloid progenitor (p) populations in the spleen. q, PCR analysis of genomic DNA from osteoblasts and indicated haematopoieic populations from wild-type and Ctnnb1CAosb mice. r–t, Real-time PCR analysis of β-catenin targets in bone marrow CD45+CD34+CD31+ cells (r), spleen (s) and bones (t). In a, n = 6; b–k, n–p, n = 8; l, m, n = 5 and r–t, n = 4 mice per group. Results are mean ± s.d. and show a representative from five (a–p) or 2 (q–t) independent experiments. *P < 0.05 versus wild type. MNC, mononuclear cells.
Extended Data Figure 2 Multi-organ infiltration with blasts and dysplastic cells and myeloid differentiation block in Ctnnb1CAosb mice.
a, Blast infiltration (solid arrows) and neutrophil hypersegmentation (open arrow and magnified panels) in the blood of Ctnnb1CAosb mice. Images at ×40 or ×100 magnification. b, Blast infiltration (solid arrows) and micro-megakaryocytes with hyperchromatic nuclei (white arrows) in the bone marrow of Ctnnb1CAosb mice. Images at ×60 magnification. c, Blast infiltration (solid arrows and magnified panel) and presence of dysplastic megakaryocytes (yellow arrows and magnified panel) in the spleen of Ctnnb1CAosb mice. Image at ×400 magnification. d–f, Myeloperoxidase (MPO) staining of long bone (d), spleen (e), and liver (f) showing massive invasion of myeloid cells. g, CD117 (c-kit) staining of bone sections showing CD117+ blasts in Ctnnb1CAosb mice. h, CD13 staining of bone sections showing myeloid/monocytic infiltration in Ctnnb1CAosb mice. In d–h images at ×60 magnification. i–l, B-cell progenitors numbers in the bone marrow (i, j), spleen (k) and lymph nodes (l). m–t, Proportion of T cells. u, Lack of myeloid cell differentiation in Ctnnb1CAosb bone marrow cells following treatment with cytokines. v–x, Percentage of immature myeloid cells in ex vivo bone marrow cultures treated with cytokines. y, Robertsonian translocation between chromosomes 1 and 19 in 2 of 30 metaphases of the spleen of an 18-day-old Ctnnb1CAosb mouse. Inset shows the same abnormality in another cell. z, Whole-exome sequencing of myeloid malignancies (CD11b+/Gr1+) from 3 Ctnnb1CAosb mice and 3 germline normal (tail) samples. In i–x, n = 6 mice per group. *P < 0.05 versus wild type. Results are mean ± s.d. and show a representative of five (i–t) or three (u–x) independent experiments.
Extended Data Figure 3 Cell autonomous AML development by bone marrow and LT-HSCs cells of Ctnnb1CAosb mice.
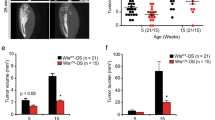
a, Engraftment efficiency of CD45.2 Ctnnb1CAosb bone marrow cells in lethally irradiated CD45.1 wild-type mice 7 weeks following transplantation. b–f, Percentage of indicated populations in bone marrow of transplanted mice. g, Blasts in blood (15–80%, solid arrows) of lethally irradiated CD45.1 wild-type mice transplanted with CD45.2 Ctnnb1CAosb marrow cells 7 weeks following transplantation. h, Blasts (solid arrows) and dysplastic megakaryocytes (open arrow) in bone marrow of transplanted mice. Images at ×60 magnification. i, Lethality curves. j, Engraftment efficiency of CD45.1 wild-type bone marrow cells in lethally irradiated CD45.2 Ctnnb1CAosb mice. k–o, Increased percentage of LSK cells (k), myeloid progenitors (l), and CD11b+/Gr1+ (m) cells and decreases in erythroid cells (n) and B-lymphopoiesis (o) in the bone marrow of transplanted mice. p, q, Blasts in the blood (p) and bone marrow (q) (black arrows) of transplanted mice. Images were taken at ×100 (o) and ×60 (p) magnification. r, Lethality curves. s, Engraftment efficiency of indicated bone marrow haematopoietic populations from 4-week-old CD45.2 Ctnnb1CAosb or wild-type mice in sublethally irradiated CD45.1 wild-type mice after 4 weeks (for LT-HSCs, due to lethality) and 8 weeks (for other populations) of transplantation. t, Blood counts in wild type mice transplanted mice. u–w, Lack of blasts in the blood of wild-type mice transplanted with indicated haematopoietic cells from Ctnnb1CAosb mice. x, Disease development in wild-type mice transplanted with indicated haematopoietic cells from Ctnnb1CAosb mice. y, Splenomegaly in wild-type mice transplanted with LT-HSCs from Ctnnb1CAosb. z, Spleen size and weight in wild-type mice transplanted with indicated haematopoietic populations from WT/ Ctnnb1CAosb mice. n = 6 mice per group. Results are mean ± s.d. and show a representative of two independent experiments. *P < 0.05 wild type–wild type versus wild type– Ctnnb1CAosb or versus Ctnnb1CAosb–wild type transplanted group.
Extended Data Figure 4 Newborn Ctnnb1CAosb mice show MDS but fetal HSCs from Ctnnb1CAosb mice do not transfer AML.
a–e, Increased percentage of LSK cells (a), GMPs (b), and CD11b+/Gr1+ (c) cells and decreases in erythroid (d) cells and B-lymphopoiesis (e) in the liver of newborn (P1) Ctnnb1CAosb mice. f–j, Increased percentage of LSK cells (f), GMPs (g), and CD11b+/Gr1+ (h) cells and decreases in erythroid cells (i) and B-lymphopoiesis (j) in the bone marrow of newborn (P1) Ctnnb1CAosb mice. k–m, Liver (k), bone marrow (l) and spleen (m) of of newborn (P1) Ctnnb1CAosb mice showing microhypolobated megakaryocytes (open arrows), Pelger Huet neutrophils (yellow arrows) or blasts (solid arrows). Images at ×100 magnification. n, Percentage of immature myeloid cells in the bone marrow of newborn mice. o–t, Flow cytometry and Giemsa-stained cytospins showing lack of changes in the percentage of immature myeloid cells in ex vivo cultures of bone marrow cells from P1 stage Ctnnb1CAosb mice and treated with indicated cytokines. u, Engraftment efficiency of CD45.2 Ctnnb1CAosb LSK cells obtained from the liver of embryonic day 18.5 embryos in sublethally irradiated CD45.1 wild-type mice. v, Normal peripheral blood measurements in transplanted mice. w, Lack of blasts in the blood of transplanted wild-type mice. Images at ×100 magnification. n = 6 mice per group. Results are mean ± s.d. and represent at least two independent experiments. *P < 0.05 versus wild type.
Extended Data Figure 5 Inhibition of increased Notch signalling normalizes blood counts and rescues haematopoietic defects in Ctnnb1CAosb mice.
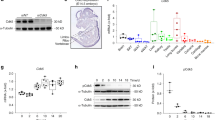
a, Microarray analysis of calvaria-derived osteoblasts from Ctnnb1CAosb mice. AML and Notch-related genes in Ctnnb1CAosb osteoblasts and with P < 0.05 and fold change of ± 20% in one comparison. Genes that are up- or downregulated relative to wild type are shown. b, Flow cytometry analysis of jagged 1expression in osteoblasts (MFI: mean fluorescent intensity). c, Luciferase activity in HEK293T cells co-transfected with β-catenin, Lef1 and Jag1-Luc reporter constructs (−4112/+130) and (−2100/+130). Results show fold induction over respective Jag1-Luc reporter constructs. *P < 0.05 versus respective Jag1-Luc. Results are mean ± s.d. d, ChIP in primary osteoblasts using anti-β-catenin antibody. Primers spanned the putative TCF/LEF binding sites (indicated) on the Jag1 promoter. e, Expression of Notch1 and Notch2 in LSK+ cells. f–g, Expression of Notch targets in LSK+ subpopulations. h, i, Normal intestinal architecture (h) and PAS staining (i) showing lack of goblet cell (arrows) metaplasia in DBZ-treated mice. Images at ×60 magnification. j, Peripheral blood counts and bone marrow cellularity in wild-type and Ctnnb1CAosb mice treated daily with vehicle or DBZ (2 µmol per kg body weight) for 10 days. k–p, Percentage of LSK cells (k), LSK+ subpopulations (l), myeloid progenitors (m), CD11b+/Gr1+ population (n), erythroid cells (o) and LSK+/FLT3+ population (p) in the bone marrow. m, Percentage of erythroid cells and p, LSK+/FLT3+ population in the bone marrow. q–s, Percentage of myeloid progenitor populations (q), CD11b+/Gr1+ (r), cells and erythroid (s), cells in the spleen. In a n = 3 mice per group and in b, n = 4 mice per group. In c, d, results represent two independent experiments. In e–g N = 4 mice per group, and *P < 0.05 versus wild type. In h–s n = 8 mice per group and *P < 0.05 versus wild type and #P < 0.05 Ctnnb1CAosb vehicle versus DBZ-treated Ctnnb1CAosb group. Results are mean ± s.d. and show a representative of two independent experiments.
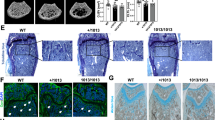
Extended Data Figure 6 Jag1 inactivation in osteoblasts prevents AML of Ctnnb1CAosb mice.
a–d, Expression of Notch transcriptional targets in bone marrow LSK subpopulations. Rescue of changes in the proportions of LSK (e) and erythroid (f) cells in the bone marrow of Ctnnb1CAosb /Jag1osb+/− mice. g, Improvement of B-lymphopoiesis in Ctnnb1CAosb;Jag1osb+/− mice. Normal (h) bone marrow, spleen (i) and liver (j) histology in Ctnnb1CAosb;Jag1osb+/− mice. k, Long bone sections. Images at ×4 magnification. In a–d, n = 4 and in e–k n = 8 mice per group. *P < 0.05 versus wild type and #P < 0.05 versus Ctnnb1CAosb;Jag1osb+/− mice. Results are mean ± s.d. and show a representative of three (a–d) and two (e–k) independent experiments.
Extended Data Figure 7 Inhibition of Notch signalling reverses AML in Ctnnb1CAosb mice.
a, Lack of blasts (solid arrows) and, normal neutrophils (right panel) in blood of DBZ-treated Ctnnb1CAosb mice. b–c, Normal megakaryocytes in the bone marrow (b) and spleen (c). d, Normal spleen histology in DBZ-treated Ctnnb1CAosb mice. Yellow arrows indicate abnormal cells with large nucleoli and dotted arrow indicates abnormal megakaryocytes in Ctnnb1CAosb mice; white arrow indicates normal megakaryocytes in DBZ-treated Ctnnb1CAosb mice. e, Lack of monocyte infiltration in the liver of DBZ-treated Ctnnb1CAosb mice. Arrow indicates cluster of mononuclear cells. f–h, MPO staining of bone marrow (f), spleen (g) and liver (h). i, Per cent of cells staining with MPO in the indicated tissues. j, Increased survival in DBZ-treated Ctnnb1CAosb mice. In a, b, images taken at ×100 magnification. In c–g images taken at ×60 magnification. k–l, Proportion of B-cell populations in the bone marrow (k) and spleen (l). m, Long bone sections. Images at ×4 magnification. n = 6 mice per group. *P < 0.05 versus wild type and #P < 0.05 Ctnnb1CAosb vehicle versus DBZ-treated Ctnnb1CAosb group. Results are mean ± s.d. and show a representative from two independent experiments.
Extended Data Figure 8 Nuclear accumulation of β-catenin in osteoblasts and increased Notch signalling in 38.3% of patients with MDS/AML and identification of underlying pre-AML conditions by nuclear localization of β-catenin in osteoblasts.
a–f, Double immunofluorescence staining with β-catenin and RUNX2 in osteoblasts from bone marrow biopsies from 6 MDS/AML patients harbouring nuclear accumulation of β-catenin in osteoblasts and showing nuclear accumulation of HEY1 in the corresponding patients (×60 magnification). g, h, During screening assumed healthy controls, 2 individuals were identified with nuclear β-catenin in their osteoblasts. Re-evaluation showed underlying hematologic disorder, Case 1: MDS RAEB-1, Case 2: Jak2 positive myelofibrosis. g, Double immunofluorescence staining with β-catenin and RUNX2 in osteoblasts from bone marrow biopsies of the 2 cases (×60 magnification). h, β-catenin cellular localization in cases 1 and 2 with associated cytogenetic abnormalities. NL: normal cytogenetics. In the fourth column percentages indicate osteoblasts with nuclear localization of β-catenin.
Extended Data Figure 9 Membrane accumulation of β-catenin in osteoblasts in 61.7% of patients with MDS/AML and in healthy subjects and nuclear accumulation of β-catenin in osteoblasts in 38.3% of patients with MDS/AML identified by flow cytometry.
Double immunofluorecsence staining with β-catenin and Runx2 in osteoblasts from bone marrow biopsies from a–c, 3 MDS/AML patients and d–g, 4 healthy subjects harbouring membrane localization of β-catenin in osteoblasts. h–j, Flow cytometry using a non-phospho β-catenin antibody detecting nuclear/activated β-catenin. Representative plots showing nuclear (h), versus non-nuclear (i), localization of β -catenin in osteoblasts from individual MDS/AML patients and, non-nuclear localization of β-catenin in osteoblasts from 5 healthy subjects as CD34−/Lin−OCN+ cells, (OCN, osteocalcin an osteoblast-specific protein used for isolation of live osteoblastic cells).
Supplementary information
Supplementary Table 1
This file shows the nuclear accumulation of β-catenin in osteoblasts of 38.29% MDS/AML patients. β-catenin cellular localization in patients with MDS, AML, and AML arising from prior MDS with associated cytogenetic abnormalities. NL: normal cytogenetics; NE: cytogenetics non-evaluable; ND: cytogenetics not determined. Percentages in parentheses in the 4th column indicate percentages of osteoblasts with nuclear localization of β-catenin; Y denotes nuclear localization of β-catenin in osteoblasts and N denotes membrane localization of β-catenin in osteoblasts. In the latter samples nuclear localization of β-catenin was not detected in any of the osteoblasts. (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Kode, A., Manavalan, J., Mosialou, I. et al. Leukaemogenesis induced by an activating β-catenin mutation in osteoblasts. Nature 506, 240–244 (2014). https://doi.org/10.1038/nature12883
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12883
This article is cited by
-
Ablation of Wnt signaling in bone marrow stromal cells overcomes microenvironment-mediated drug resistance in acute myeloid leukemia
Scientific Reports (2024)
-
The role of bone marrow microenvironment (BMM) cells in acute myeloid leukemia (AML) progression: immune checkpoints, metabolic checkpoints, and signaling pathways
Cell Communication and Signaling (2023)
-
The roles of bone remodeling in normal hematopoiesis and age-related hematological malignancies
Bone Research (2023)
-
MacroH2A1.1 as a crossroad between epigenetics, inflammation and metabolism of mesenchymal stromal cells in myelodysplastic syndromes
Cell Death & Disease (2023)
-
A mysterious triangle of blood, bones, and nerves
Journal of Bone and Mineral Metabolism (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.