Abstract
Strigolactones (SLs) are a group of newly identified plant hormones that control plant shoot branching. SL signalling requires the hormone-dependent interaction of DWARF 14 (D14), a probable candidate SL receptor, with DWARF 3 (D3), an F-box component of the Skp–Cullin–F-box (SCF) E3 ubiquitin ligase complex. Here we report the characterization of a dominant SL-insensitive rice (Oryza sativa) mutant dwarf 53 (d53) and the cloning of D53, which encodes a substrate of the SCFD3 ubiquitination complex and functions as a repressor of SL signalling. Treatments with GR24, a synthetic SL analogue, cause D53 degradation via the proteasome in a manner that requires D14 and the SCFD3 ubiquitin ligase, whereas the dominant form of D53 is resistant to SL-mediated degradation. Moreover, D53 can interact with transcriptional co-repressors known as TOPLESS-RELATED PROTEINS. Our results suggest a model of SL signalling that involves SL-dependent degradation of the D53 repressor mediated by the D14–D3 complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cook, C. E., Whichard, L. P., Turner, B., Wall, M. E. & Egley, G. H. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190 (1966)
Akiyama, K., Matsuzaki, K. & Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 (2005)
Gomez-Roldan, V. et al. Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008)
Umehara, M. et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 (2008)
Domagalska, M. A. & Leyser, O. Signal integration in the control of shoot branching. Nature Rev. Mol. Cell Biol. 12, 211–221 (2011)
Ruyter-Spira, C., Al-Babili, S., van der Krol, S. & Bouwmeester, H. The biology of strigolactones. Trends Plant Sci. 18, 72–83 (2013)
Stirnberg, P., van De Sande, K. & Leyser, O. MAX1 and MAX2 control shoot lateral branching in Arabidopsis . Development 129, 1131–1141 (2002)
Sorefan, K. et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17, 1469–1474 (2003)
Booker, J. et al. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14, 1232–1238 (2004)
Booker, J. et al. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8, 443–449 (2005)
Stirnberg, P., Furner, I. J. & Leyser, O. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94 (2007)
Waters, M. T., Brewer, P. B., Bussell, J. D., Smith, S. M. & Beveridge, C. A. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 159, 1073–1085 (2012)
Waters, M. T. et al. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis . Development 139, 1285–1295 (2012)
Ishikawa, S. et al. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46, 79–86 (2005)
Zou, J. et al. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48, 687–698 (2006)
Arite, T. et al. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029 (2007)
Lin, H. et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21, 1512–1525 (2009)
Arite, T. et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424 (2009)
Gao, Z. et al. Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol. Biol. 71, 265–276 (2009)
Liu, W. et al. Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230, 649–658 (2009)
Johnson, X. et al. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 142, 1014–1026 (2006)
Snowden, K. C. et al. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17, 746–759 (2005)
Simons, J. L., Napoli, C. A., Janssen, B. J., Plummer, K. M. & Snowden, K. C. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol. 143, 697–706 (2007)
Hamiaux, C. et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 (2012)
Wang, Y. & Li, J. Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279 (2008)
Alder, A. et al. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351 (2012)
Zhao, L. H. et al. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439 (2013)
Kagiyama, M. et al. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160 (2013)
Smith, S. M. & Waters, M. T. Strigolactones: destruction-dependent perception? Curr. Biol. 22, R924–R927 (2012)
Iwata, N. & Omura, T. Studies on the trisomics in rice plants (Oryza sativa L.). VI. An accomplishment of a trisomic series in japonica rice plants. Jpn. J. Genet. 59, 199–204 (1984)
Wei, L. R., Xu, J. C., Li, X. B., Qian, Q. & Zhu, L. H. Genetic analysis and mapping of the dominant dwarfing dene D-53 in rice. J. Integr. Plant Biol. 48, 447–452 (2006)
Stanga, J. P., Smith, S. M., Briggs, W. R. & Nelson, D. C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis . Plant Physiol. 163, 318–330 (2013)
Szemenyei, H., Hannon, M. & Long, J. A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008)
Pauwels, L. et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010)
Fukui, K. et al. New branching inhibitors and their potential as strigolactone mimics in rice. Bioorg. Med. Chem. Lett. 21, 4905–4908 (2011)
Fukui, K., Ito, S. & Asami, T. Selective mimics of strigolactone actions and their potential use for controlling damage caused by root parasitic weeds. Mol. Plant 6, 88–99 (2013)
Nelson, D. C. et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana . Proc. Natl Acad. Sci. USA 108, 8897–8902 (2011)
Causier, B., Ashworth, M., Guo, W. & Davies, B. The TOPLESS interactome: a framework for gene repression in Arabidopsis . Plant Physiol. 158, 423–438 (2012)
Long, J. A., Ohno, C., Smith, Z. R. & Meyerowitz, E. M. TOPLESS regulates apical embryonic fate in Arabidopsis . Science 312, 1520–1523 (2006)
Yoshida, A., Ohmori, Y., Kitano, H., Taguchi-Shiobara, F. & Hirano, H. Y. Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 70, 327–339 (2012)
Kwon, Y. et al. OsREL2, a rice TOPLESS homolog functions in axillary meristem development in rice inflorescence. Plant Biotechnol. Rep. 6, 213–224 (2012)
Gao, X. et al. OsLIS-L1 encoding a lissencephaly type-1-like protein with WD40 repeats is required for plant height and male gametophyte formation in rice. Planta 235, 713–727 (2012)
Iwata, N., Satoh, N. & Omura, T. Linkage studies in rice. Linkage groups for 6 genes newly described. Jpn. J. Breed. 27, (suppl.), 250–251 1977)
Wang, Z. et al. A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22, 409–417 (2004)
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994)
Bart, R., Chern, M., Park, C. J., Bartley, L. & Ronald, P. C. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2, 13 (2006)
Kierzkowski, D. et al. The Arabidopsis CBP20 targets the cap-binding complex to the nucleus, and is stabilized by CBP80. Plant J. 59, 814–825 (2009)
Kamachi, K., Yamaya, T., Mae, T. & Ojima, K. A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiol. 96, 411–417 (1991)
Chu, C. C. et al. Establishment of an efficient medium for anther culture of rice through comparative experiments on nitrogen-source. Sci. China 18, 659–668 (1975)
Gamborg, O. L., Miller, R. A. & Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158 (1968)
Acknowledgements
We thank K. Yoneyama for assistance in SL analysis and C. Yan for SL measurement. This work was supported by grants from Ministry of Science and Technology of the People’s Republic of China (2012AA10A301) and National Natural Science Foundation of China (31025004, 90817108 and 91217311).
Author information
Authors and Affiliations
Contributions
L.J., X.L., G.X. and H.L. designed research, performed experiments, analysed data and wrote the paper; F.C., L.W., X.M., H.Y., Y.Y., G.L., W.Y., L.Z., H.M., Y.H., Z.W. and K.M. performed some of the experiments. Q.Q. provided and planted the rice materials. J.L., Y.W. and H.E.X. designed research, analysed data and wrote the paper. J.L. conceived and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Characterization of the e9 mutant.
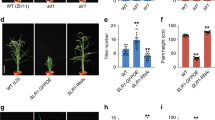
a, Tiller number and plant height of wild-type, homozygous and heterozygous e9 plants at the heading stage. Values are means ± s.d. (n = 8). The double asterisks represent significant difference determined by the Student’s t-test at P < 0.01. b, Kinetic comparison of tiller numbers between wild-type and e9 plants at different developmental stages. Values are means ± s.d. (n = 8). DAG, days after germination. Rice plants (a and b) were cultivated in the field in Beijing in the natural growing season. c, Four-week-old seedlings of wild-type, e9, d3 and d27 upon 1 μM GR24 treatment. Scale bars, 10 cm. Ten individual plants for each material were treated with GR24. d, The expression levels of the D10 gene revealed by qPCR in wild-type, e9, d3 and d27 mutant seedlings. Values are means ± s.d. (n = 3). The double asterisks represent significant difference determined by the Student’s t-test at P < 0.01. e, The representative LC/MS–MS chromatograms for epi-5DS in root exudates of wild type and e9 before calibration against an internal standard.
Extended Data Figure 2 Cloning and confirmation of E9/D53.
a, E9 was mapped in the interval between molecular markers Ds3 and K81114 on chromosome 11 using 142 recessive individual plants showing normal tillering phenotype from an F2 population. Numbers under the markers indicate recombinants. b, E9/D53 mutation sites in e9/d53 in the coding region and its amino acid changes. c, Schematic diagram of D53:d53 constructs. d, Phenotypes of wild-type and D53:d53 transgenic plants at the mature stage. Scale bar, 20 cm. Nine independent transgenic lines showed the similar tillering and dwarf phenotypes. e, Comparison of tiller numbers between wild-type and D53:d53 transgenic plants at the mature stage. Values are means ± s.d. (n = 15). The double asterisks represent significant difference determined by the Student’s t-test at P < 0.01. The plants were grown in the field in Beijing in the natural growing season.
Extended Data Figure 3 Phylogenetic tree of D53-like family proteins in rice and Arabidopsis.
BlastP was done using the D53 protein sequence against all rice and Arabidopsis proteins at the MSU Rice Genome Annotation Project. In the BlastP result, rice proteins were filtered using cut-off E value < 0.1 plus top query coverage > 10% and Arabidopsis proteins were filtered using cut-off E value < 0.0005. Multiple sequence alignment of the protein sequences was done using Clustalw2. Maximum-likelihood phylogenetic tree was drawn by MEGA 5.05 using default parameters with 100 times bootstrapping. Numbers above the branches represent bootstrap support based on 100 bootstrap replicates. Branch length represents substitutions per site.
Extended Data Figure 4 Alignment of D53 and D53-like proteins in rice and Arabidopsis.
The graphic view of alignment was generated by BioEdit using Clustalw for multiple sequence alignment of protein sequences. Blue underline refers to the Double Clp-N domain, light green to atypical walker A motifs, dark green box to walker B motifs and red boxes to putative EAR motifs. D53, Os11g01330; D53-like, Os12g01360; AtD53-like 1, At1g07200; AtD53-like 2, At2g29970; AtD53-like 3, At2g40130; AtHSP101, At1g74310; OsHSP101, Os05g44340.
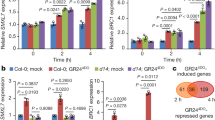
Extended Data Figure 5 D53 expression levels in d mutants and the mutation sites in other d mutants used in this study.
a, Expression levels of D53 revealed by qPCR in wild-type, d3, d10, d14, d17 and d27 seedlings. Values are means with s.d. of three independent experiments. The double asterisks represent significant difference determined by the Student’s t-test at P < 0.01. b, The mutation sites in other d mutants identified in this study.
Extended Data Figure 6 D3 and D14 protein levels are unaffected by GR24 treatment.
a, Phenotypes of mature transgenic plants of D3-GFP and D14-GFP in the d3 or d14 background, respectively, showing that D3–GFP and D14–GFP can rescue the corresponding phenotypes of d3 and d14. Scale bar, 10 cm. Five independent transgenic lines were shown to have similar phenotypes. b, Comparison of the tiller number of transgenic plants of D3-GFP/d3 and D14-GFP/d14. Values are means ± s.d. (n = 5). The double asterisks represent significant difference determined by the Student’s t-test at P < 0.01. c, D3–GFP abundance in d3 treated with 10 μM GR24 for 60 min. Protein level was analysed by immunoblotting using GFP antibody. d, The D14–GFP protein level in the d14 background as analysed by immunoblotting using GFP antibody. Transgenic plants of D14-GFP in the d14 background were treated with 10 μM GR24 for 60 min. Rice plants (T2 generation) were grown in the field in Hainan province.
Extended Data Figure 7 Effects of GR24 and SL mimics on rice tillering.
a, Chemical structures of GR24 and SL mimics. A, 4-[(2,5-dihydro-4-methyl-5-oxo-2-furanyl)oxy]benzonitrile; B, 5-(4-bromophenoxy)-3-methyl-2(5H)-furanone; C, 5-(4-iodophenoxy)-3-methyl-2(5H)-furanone. D, 3,3a,4,8b-tetrahydro-3-(hydroxymethylene)-2H-Indeno[1,2-b]furan-2-one (ABC rings of GR24); E, 5-hydroxy-3-methyl-2(5H)-furanone (D ring of GR24). b, Effects of 1 μM GR24 and SL mimics on tillering of 4-week-old seedlings. Values are means ± s.d. (n = 5).
Extended Data Figure 8 Phenotypes of transgenic plants of constitutively expressed D53–GFP in d3 and d14.
a, Phenotypes of wild-type, d3 and Act:D53-GFP/d3 transgenic plants at the mature stage. Scale bar, 10 cm. Six independent transgenic lines showed similar phenotypes. b, Phenotypes of wild-type, d14 and Act:D53-GFP/d14 transgenic plants at the mature stage. Scale bar, 10 cm. Six independent transgenic plants showed similar phenotypes. c, Tiller numbers of Act: D53-GFP/d14 and Act:D53-GFP/d3 transgenic plants at the mature stage. Values are means ± s.d. (n = 8). d, The D53–GFP abundance in d3 or d14 plants treated with 10 μM GR24 for 60 min. Protein levels were analysed by immunoblotting using GFP antibody. Rice plants (T1 generation) were grown in the field in Hainan province.
Extended Data Figure 9 Phenotypes and confirmation of transgenic plants of D53-RNAi in d3 and d14.
a, Phenotypes of wild-type, d3 and D53-RNAi/d3 transgenic plants. Scale bar, 10 cm. Four independent transgenic plants showed similar phenotypes. b, Phenotypes of wild-type, d14 and D53-RNAi/d14 transgenic plants at the mature stage. Scale bar, 10 cm. Five independent transgenic plants showed similar phenotypes. c, Tiller numbers of transgenic plants of D53-RNAi in d3 and d14 backgrounds at the mature stage. Values are means ± s.d. (n = 5). The double asterisks represent significant difference determined by the Student’s t-test at P < 0.01. d, The D53 abundance of D53-RNAi in d3 or d14 transgenic plants. Protein levels were analysed by immunoblotting using D53 polyclonal antibodies. Rice plants (T1 generation) were cultivated in the field in Hainan province. e, Sequence information of D53-RNAi constructs. Two complementary inverted D53 fragments are shown in black and the intron sequence between inverted DNA fragments is in red.
Extended Data Figure 10 Determination of D53 antibody specificity.
Determination of D53 antibody specificity by D53–GFP transgenic calli. Left, anti-GFP (Roche), 1:10,000 dilution; right, anti-D53 (1:10,000). Horseradish-peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG antibodies were used as secondary antibodies to detect anti-D53 and anti-GFP.
Supplementary information
Supplementary Tables
This fie contains Supplementary Tables 1-2. (PDF 135 kb)
Rights and permissions
About this article
Cite this article
Jiang, L., Liu, X., Xiong, G. et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 (2013). https://doi.org/10.1038/nature12870
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12870
This article is cited by
-
Strigolactone regulates nitrogen-phosphorus balance in rice
Science China Life Sciences (2024)
-
A mutation in CsDWF7 gene encoding a delta7 sterol C-5(6) desaturase leads to the phenotype of super compact in cucumber (Cucumis sativus L.)
Theoretical and Applied Genetics (2024)
-
Strigolactone and abscisic acid synthesis and signaling pathways are enhanced in the wheat oligo-tillering mutant ot1
Molecular Breeding (2024)
-
Genome-wide identification and analysis of the SUPPRESSOR of MAX2 1-LIKE gene family and its interaction with DWARF14 in poplar
BMC Plant Biology (2023)
-
FRIZZLE PANICLE (FZP) regulates rice spikelets development through modulating cytokinin metabolism
BMC Plant Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.