Abstract
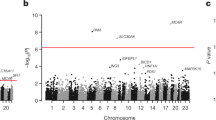
Performing genetic studies in multiple human populations can identify disease risk alleles that are common in one population but rare in others1, with the potential to illuminate pathophysiology, health disparities, and the population genetic origins of disease alleles. Here we analysed 9.2 million single nucleotide polymorphisms (SNPs) in each of 8,214 Mexicans and other Latin Americans: 3,848 with type 2 diabetes and 4,366 non-diabetic controls. In addition to replicating previous findings2,3,4, we identified a novel locus associated with type 2 diabetes at genome-wide significance spanning the solute carriers SLC16A11 and SLC16A13 (P = 3.9 × 10−13; odds ratio (OR) = 1.29). The association was stronger in younger, leaner people with type 2 diabetes, and replicated in independent samples (P = 1.1 × 10−4; OR = 1.20). The risk haplotype carries four amino acid substitutions, all in SLC16A11; it is present at ∼50% frequency in Native American samples and ∼10% in east Asian, but is rare in European and African samples. Analysis of an archaic genome sequence indicated that the risk haplotype introgressed into modern humans via admixture with Neanderthals. The SLC16A11 messenger RNA is expressed in liver, and V5-tagged SLC16A11 protein localizes to the endoplasmic reticulum. Expression of SLC16A11 in heterologous cells alters lipid metabolism, most notably causing an increase in intracellular triacylglycerol levels. Despite type 2 diabetes having been well studied by genome-wide association studies in other populations, analysis in Mexican and Latin American individuals identified SLC16A11 as a novel candidate gene for type 2 diabetes with a possible role in triacylglycerol metabolism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Data deposits
Genotype data have been deposited in dbGaP under accession number phs000683.v1.p1. Microarray data used in the ‘55k screen’ is publicly available through the NCBI Gene Expression Omnibus and the Cancer Cell Line Encyclopedia. A list of sample identities and accession numbers are available in the Supplementary Information.
References
Rosenberg, N. A. et al. Genome-wide association studies in diverse populations. Nature Rev. Genet. 11, 356–366 (2010)
Grant, S. F. A. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genet. 38, 320–323 (2006)
Unoki, H. et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nature Genet. 40, 1098–1102 (2008)
Yasuda, K. et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nature Genet. 40, 1092–1097 (2008)
Villalpando, S. et al. Prevalence and distribution of type 2 diabetes mellitus in Mexican adult population: a probabilistic survey. Salud Publica Mex. 52, S19–S26 (2010)
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006)
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009)
Williams, A. L., Patterson, N., Glessner, J., Hakonarson, H. & Reich, D. Phasing of many thousands of genotyped samples. Am. J. Hum. Genet. 91, 238–251 (2012)
The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Zaitlen, N. et al. Informed conditioning on clinical covariates increases power in case-control aAssociation Studies. PLoS Genet. 8 e1003032 http://dx.doi.org/10.1371/journal.pgen.1003032 (2012)
Voight, B. F. et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genet. 42, 579–589 (2010)
Halestrap, A. P. The monocarboxylate transporter family—Structure and functional characterization. IUBMB Life 64, 1–9 (2012)
Ng, P. C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003)
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010)
Meyer, M. et al. A high-coverage genome sequence from an Archaic Denisovan individual. Science 338, 222–226 (2012)
Mednikova, M. B. A proximal pedal phalanx of a Paleolithic hominin from denisova cave, Altai. Archaeol. Ethnol. Anthropol. Eurasia 39, 129–138 (2011)
Max Planck Institute for Evolutionary Anthropology. A high-quality Neandertal genome sequence. http://www.eva.mpg.de/neandertal/ (2013)
Hublin, J. J. The origin of Neandertals. Proc. Natl Acad. Sci. USA 106, 16022–16027 (2009)
Halestrap, A. P. & Wilson, M. C. The monocarboxylate transporter family—Role and regulation. IUBMB Life 64, 109–119 (2012)
Garcia, C. K., Goldstein, J. L., Pathak, R. K., Anderson, R. G. W. & Brown, M. S. Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: Implications for the Cori cycle. Cell 76, 865–873 (1994)
Fu, S., Watkins, S. M. & Hotamisligil, G. S. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 15, 623–634 (2012)
Rhee, E. P. et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 121, 1402–1411 (2011)
Savage, D. B. & Semple, R. K. Recent insights into fatty liver, metabolic dyslipidaemia and their links to insulin resistance. Curr. Opin. Lipidol. 21, 329–336 (2010)
Samuel, V. T. & Shulman, G. I. Mechanisms for insulin resistance: Common threads and missing links. Cell 148, 852–871 (2012)
Florez, J. et al. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia 52, 1528–1536 (2009)
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009)
Risch, N. & Merikangas, K. The future of genetic studies of complex human diseases. Science. 273, 1516–1517 (1996)
Hara, K. et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum. Mol. Genet. http://dx.doi.org/10.1093/hmg/ddt399 (14 August 2013)
Acknowledgements
We thank M. Daly, V. Mootha, E. Lander and K. Estrada for comments on the manuscript, B. Voight, A. Segre, J. Pickrell and the Scientific Advisory Board of the SIGMA Project (especially C. Bustamante) for useful discussions, and A. Subramanian and V. Rusu for assistance with expression analyses. This work was conducted as part of the Slim Initiative for Genomic Medicine, a joint US–Mexico project funded by the Carlos Slim Health Institute. The UNAM/INCMNSZ Diabetes Study was supported by Consejo Nacional de Ciencia y Tecnología grants 138826, 128877, CONACyT- SALUD 2009-01-115250, and a grant from Dirección General de Asuntos del Personal Académico, UNAM, IT 214711. The Diabetes in Mexico Study was supported by Consejo Nacional de Ciencia y Tecnología grant 86867 and by Instituto Carlos Slim de la Salud, A.C. The Mexico City Diabetes Study was supported by National Institutes of Health (NIH) grant R01HL24799 and by the Consejo Nacional de Ciencia y Tenologia grants 2092, M9303, F677-M9407, 251M and 2005-C01-14502, SALUD 2010-2-151165. The Multiethnic Cohort was supported by NIH grants CA164973, CA054281 and CA063464. The Singapore Chinese Health Study was funded by the National Medical Research Council of Singapore under its individual research grant scheme and by NIH grants R01 CA55069, R35 CA53890, R01 CA80205 and R01 CA144034. The Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) project was supported by NIH grant U01DK085526. The San Antonio Mexican American Family Studies (SAMAFS) were supported by R01 DK042273, R01 DK047482, R01 DK053889, R01 DK057295, P01 HL045522 and a Veterans Administration Epidemiologic grant to R.A.D. A.L.W. was supported by National Institutes of Health Ruth L. Kirschstein National Research Service Award number F32 HG005944.
Author information
Authors and Affiliations
Consortia
Contributions
See the author list for details of author contributions.
A list of participants and affiliations for the T2D-GENES Consortium and the Broad Genomics Platform is available in the Supplementary Information.
Corresponding authors
Ethics declarations
Competing interests
The author declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Principal component analysis (PCA) projection of SIGMA samples onto principal components calculated using data from samples collected by the Human Genome Diversity Project (HGDP) and 1000 Genomes Project.
a, b, PCA projection of SIGMA onto HGDP Yoruba, French, Karitiana and Han (Chinese) populations before ancestry quality control filters were applied (a), with cohort centroids as indicated, and after all quality control filters were applied (b), with case and control centroids as indicated. c, d, Principal components 3 and 4 before filtering samples on ancestry (a small number of samples in the MEC show East Asian admixture) (c), and after all quality control filters were applied (d). e, f, Additional plots as in b but separating cases (e) and controls (f). g, SIGMA samples projected onto the 1000 Genomes Project Omni2.5 genotype data. 1000 Genomes samples are labelled by their continental ancestry group: AFR, African; AMR, Native American descent; ASN, east Asian; EUR, European.
Extended Data Figure 2 Regional plot for signal at TCF7L2.
Point colour indicates r2 to the most strongly associated site (rs7903146) and recombination rate is also shown, both based on the 1000 Genomes ASN population.
Extended Data Figure 3 Conditional analyses reveal multiple independent signals at INS–IGF2 and KCNQ1.
a–d, Regional plots are shown for the interval spanning INS–IGF2 and KCNQ1 without conditioning (a), conditional on rs2237897 at KCNQ1 (b), conditional on rs2237897 and rs139647931 (both at KCNQ1) (c), and conditional on rs2237897 and rs139647931 (both at KCNQ1), and rs11564732 (the top associated variant in the INS–IGF2–TH region) (d). The top SNPs in 11p15.5 and KCNQ1 are ∼700 kb away from each other, but despite this proximity, there is a strong residual signal of association at INS–IGF2 after analysis conditional on genotype at KCNQ1. Point colour indicates r2 to rs11564732 and recombination rate is also shown, both based on the 1000 Genomes ASN population.
Extended Data Figure 4 Regional plots for SLC16A11 conditional on associated missense variants of that gene.
a–e, Association signal at chromosome 17p13 without conditioning (a), or conditional on the four missense SNPs in SLC16A11: rs117767867 (b), rs13342692 (c), rs75418188 (d) and rs75493593 (e). Point colour indicates r2 to the most strongly associated SNP (rs13342232) and recombination rate is also shown, both based on the 1000 Genomes ASN population.
Extended Data Figure 5 Cases with risk haplotype develop type 2 diabetes younger and at a lower BMI than non-carriers.
a, Distribution of age-of-onset in type 2 diabetes cases based on genotype at rs13342232, binned every 5 years with upper bounds indicated (carriers n = 1,126; non-carriers n = 594). b, Distribution of BMI in type 2 diabetes cases for carriers and non-carriers of rs13342232, binned every 2.5 kg m−2 with upper bounds indicated (carriers n = 2,161; non-carriers n = 1,647). P values from two-sample t-test between type 2 diabetes risk haplotype carriers and type 2 diabetes non-carriers.
Extended Data Figure 6 Frequency distribution of the risk haplotype and dendrogram depicting clustering with Neanderthal haplotypes.
a, Allele frequency of missense SNP rs117767867 (tag for risk haplotype) in the 1000 Genomes Phase I data set. b, Dendrogram generated from haplotypes across the 73-kb Neanderthal introgressed region. Nodes for modern human haplotypes are labelled in red or blue with the 1000 Genomes population in which the corresponding haplotype resides. Archaic Neanderthal sequences are labelled in black and include the low-coverage Neanderthal sequence14 (labelled Vindija), and the unpublished Neanderthal sequence that is homozygous for the 5 SNP risk haplotype17 (Altai). H1 includes haplotypes from MXL and FIN, and H2 and H3 both include haplotypes from CLM, MXL, CHB and ASW. Modern human sequences included are all 1000 Genomes Phase I samples that are homozygous for the 5 SNP risk haplotype (n = 15), and 16 non-risk haplotypes—four haplotypes (from two randomly selected individuals) from each of the CLM (Colombian in Medellin, Colombia), MXL (Mexican Ancestry in Los Angeles, California), CHB (Han Chinese in Beijing, China) and FIN (Finnish in Finland) 1000 Genomes populations (the populations with carriers of the 5 SNP haplotype). The red subtree depicts the Neanderthal clade, with all risk haplotypes clustering with the Altai and Vindija sequences. In blue are all other modern human haplotypes. The dendrogram was generated by the R function hclust using a complete linkage clustering algorithm on a distance matrix measuring the fraction of SNPs called in the 1000 Genomes project at which a pair of haplotypes differs (the y axis represents this distance). Because haplotypes are unavailable for the archaic samples, we picked a random allele to compute the distance matrix.
Extended Data Figure 7 Analysis of gene expression for SLC16A11, SLC16A13 and SLC16A1 in 30 human tissues.
Data measured using nCounter are shown as mean, normalized mRNA counts per 200 ng RNA ± s.e.m. Threshold for background (nonspecific) binding is indicated by the red line. Sample size for each tissue (n): pancreas (5); adipose, brain, colon, liver, skeletal muscle and thyroid (3); adrenal, fetal brain, breast, heart, kidney, lung, placenta, prostate, small intestine, spleen, testes, thymus and trachea (2); bladder, cervix, oesophagus, fetal liver, ovary, salivary gland, fetal skeletal muscle, skin, umbilical cord and uterus (1).
Extended Data Figure 8 Microarray-based analysis of SLC16A11 expression in human tissues.
a, Results from the ‘55k screen’, a survey of gene expression in 55,269 samples profiled on the Affymetrix U133 plus 2.0 array, are shown as the fraction of samples of a given tissue in which SLC16A11 is expressed. Sample size for each tissue (n): adipose (394), adrenal (69), brain (1,990), breast (4,104), heart (178), kidney (675), liver (721), lung (1,442), pancreas (150), placenta (107), prostate (578), salivary gland (26), skeletal muscle (793), skin (947), testis (102), thyroid (108). b, Histograms show the expression level distribution of SLC16A11 and other well-studied liver genes in 721 liver samples from the ‘55k screen’. INS is shown as reference for a gene not expressed in liver. On the basis of negative controls, a normalized log2 expression of 4 is considered baseline and log2 expression values greater than 6 are considered expressed.
Extended Data Figure 9 SLC16A13 localizes to Golgi apparatus.
a, b, HeLa cells transiently expressing C terminus, V5-tagged SLC16A13 (a) or BFP (b) were immunostained for SLC16A13 or BFP expression (anti-V5) along with specific markers for the endoplasmic reticulum (anti-calnexin), cis-Golgi apparatus (anti-Golph4) and mitochondria (MitoTracker). Representative images from multiple independent transfections are shown. Owing to heterogeneity in expression levels of overexpressed proteins and endogenous organelle markers, imaging of each protein was optimized for clarity of localization and varied across images; therefore, images are not representative of relative expression levels of each protein as compared to the other proteins.
Extended Data Figure 10 Pathway and class-based metabolic changes induced by SLC16A11 expression.
Changes in metabolite levels in HeLa cells expressing SLC16A11–V5 compared to control-transfected cells are plotted in groups according to metabolic pathway or class. Data shown are the combined results from three independent experiments, each of which included 12 biological replicates each for SLC16A11 and control. Pathways shown include all KEGG pathways from the human reference set for which metabolites were measured as well as eight additional classes of metabolites covering carnitines and lipid subtypes. Each point within a pathway or class shows the fold change of a single metabolite within that pathway or class. For each pathway or class with at least six measured metabolites, enrichment was computed as described in Supplementary Methods. Asterisks indicate pathways with P ≤ 0.05 and FDR ≤ 0.25. Supplementary Table 14 shows additional details from the enrichment analysis.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Tables 1-14, Supplementary Notes, list of Subconsortia Authors and additional references. (PDF 760 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
The SIGMA Type 2 Diabetes Consortium. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506, 97–101 (2014). https://doi.org/10.1038/nature12828
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12828
This article is cited by
-
Ancestry-driven metabolite variation provides insights into disease states in admixed populations
Genome Medicine (2023)
-
Leveraging fine-scale population structures for precision healthcare
Nature Medicine (2023)
-
Sex differences in the influence of type 2 diabetes (T2D)-related genes, parental history of T2D, and obesity on T2D development: a case–control study
Biology of Sex Differences (2023)
-
Association of tyrosine hydroxylase 01 (TH01) microsatellite and insulin gene (INS) variable number of tandem repeat (VNTR) with type 2 diabetes and fasting insulin secretion in Mexican population
Journal of Endocrinological Investigation (2023)
-
The power of TOPMed imputation for the discovery of Latino-enriched rare variants associated with type 2 diabetes
Diabetologia (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.