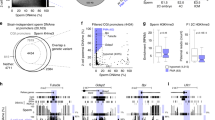
Abstract
Genomic imprinting is an allele-specific gene expression system that is important for mammalian development and function1. The molecular basis of genomic imprinting is allele-specific DNA methylation1,2. Although it is well known that the de novo DNA methyltransferases Dnmt3a and Dnmt3b are responsible for the establishment of genomic imprinting3, how the methylation mark is erased during primordial germ cell (PGC) reprogramming remains unclear. Tet1 is one of the ten-eleven translocation family proteins, which have the capacity to oxidize 5-methylcytosine (5mC)4,5,6, specifically expressed in reprogramming PGCs7. Here we report that Tet1 has a critical role in the erasure of genomic imprinting. We show that despite their identical genotype, progenies derived from mating between Tet1 knockout males and wild-Peg10 and Peg3, which exhibit aberrant hypermethylation in the paternal allele of differential methylated regions (DMRs). RNA-seq reveals extensive dysregulation of imprinted genes in the next generation due to paternal loss of Tet1 function. Genome-wide DNA methylation analysis of embryonic day 13.5 PGCs and sperm of Tet1 knockout mice revealed hypermethylation of DMRs of imprinted genes in sperm, which can be traced back to PGCs. Analysis of the DNA methylation dynamics in reprogramming PGCs indicates that Tet1 functions to wipe out remaining methylation, including imprinted genes, at the late reprogramming stage. Furthermore, we provide evidence supporting the role of Tet1 in the erasure of paternal imprints in the female germ line. Thus, our study establishes a critical function of Tet1 in the erasure of genomic imprinting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bartolomei, M. S. & Ferguson-Smith, A. C. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 3, a002592 (2011)
Li, Y. & Sasaki, H. Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res. 21, 466–473 (2011)
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004)
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010)
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009)
Shen, L. & Zhang, Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr. Opin. Cell Biol. 25, 289–296 (2013)
Yamaguchi, S. et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature 492, 443–447 (2012)
Nelissen, E. C. M., van Montfoort, A. P. A., Dumoulin, J. C. M. & Evers, J. L. H. Epigenetics and the placenta. Hum. Reprod. Update 17, 397–417 (2011)
Ono, R. et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nature Genet. 38, 101–106 (2006)
Ono, R. et al. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 13, 1696–1705 (2003)
Hackett, J. A. et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452 (2013)
Kagiwada, S., Kurimoto, K., Hirota, T., Yamaji, M. & Saitou, M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 32, 340–353 (2013)
Yamaguchi, S. et al. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23, 329–339 (2013)
Seisenberger, S. et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 (2012)
Kobayashi, H. et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 23, 616–662 (2013)
Dawlaty, M. M. et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323 (2013)
Dawlaty, M. M. et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 (2011)
Cortellino, S. et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 (2011)
Popp, C. et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463, 1101–1105 (2010)
Piccolo, F. M. et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol. Cell 49, 1023–1033 (2013)
Kobayashi, H. et al. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 113, 130–137 (2006)
Tomizawa, S.-i. et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138, 811–820 (2011)
Gu, H. et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protocols 6, 468–481 (2011)
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578 (2012)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Boyle, P. et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 13, R92 (2012)
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011)
Illingworth, R. S. et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 6, e1001134 (2010)
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnol. 28, 495–501 (2010)
Blake, J. A. et al. The mouse genome database genotypes:phenotypes. Nucleic Acids Res. 37, D712–D719 (2009)
Acknowledgements
We thank W. Jiang for help with FACS sorting of PGCs; A. Inoue, S. Matoba and D. Cai for critical reading of the manuscript. This project is supported by NIH U01DK089565 (to Y.Z.). S.Y. is supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science (JSPS). Y.Z. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
S.Y. and Y.Z. conceived the project, designed the experiments and wrote the manuscript; S.Y., L.S., Y.L. and D.S. performed experiments and analysed the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Tet1 paternal knockout mice exhibit various phenotypes, including fetal and postnatal growth defects, and neonatal and embryonic lethality.
a, Scatter plot of body and placental sizes for each pup. b, Average placental weight of E13.5 and E16.5 embryos. Error bars indicate s.e.m. **P < 0.01. c, Percentage of pups that die within 3 days of birth. **P < 0.01. d, Box plot presentation of body weights of 4- and 8-week-old control and paternalKO mice. Note that paternalKO mice exhibited a larger variation and smaller body weight compared to the control mice. Middle lines in the coloured boxes indicate the median, box edges indicate the 25th/75th percentiles, and whiskers the 0th/100th percentiles. e, Mating scores of Tet1+/− male × wild-type female (×Het) and Tet1−/− male × wild-type female (×KO) mice. Black dots indicate the numbers of pups for each litter recovered by Caesarean section at E19.5. Red lines indicate the mean litter size. f, The embryos and placentae at E13.5 recovered from four litters of Tet1 paternalKO mice. The dotted ovals indicate the residues of absorbed embryos. g, E10.5 paternalKO embryos recovered from five litters. Dotted oval indicates morphologically abnormal embryos. h, Top: E9.5 paternalKO embryos recovered from two litters. Dotted ovals indicate small embryos whose placentae are morphologically abnormal. Bottom: placentae corresponding to the embryos on the top panels. Red rectangles indicate morphologically abnormal placentae. i, Representative images of haematoxylin and eosin staining of E9.5 placentae. Red rectangles indicate the enlarged regions shown at the right panels. Ch, chorionic plate; Gi, trophoblast giant cells; La, labyrinthine zone; Sp, spongioblast. j, Schematic representation of Tet1 paternalKO phenotypes. Each bar represents an individual mouse. About 40% of paternalKO mice exhibit early embryonic lethality. About half of the remaining mice die neonatally, and about half of the surviving mice show postnatal growth defects. In contrast, abnormality is rarely observed in control, except at the neonatal stage (less than 10%).
Extended Data Figure 2 Dysregulation of imprinted genes in Tet1 paternal knockout embryos.
a, Relative expression levels of imprinted genes in E9.5 embryos analysed by RNA-seq. The averaged FPKM (fragments per kb of exon per million fragments mapped) value of three control embryos is set as 1. Arrows indicate dysregulated embryos. The location of each gene is indicated in the diagram on top of the panel. b, Relative average expression change of maternally and paternally imprinted genes. The imprinted genes that are highly expressed in E9.5 embryos (FPKM >1.0) are used in this analysis. Error bars indicate s.e.m. *P < 0.05. c, Summary of imprinted genes dysregulated in paternalKO embryos. N, normal size; S, small size; +, normal expression; −, downregulated.
Extended Data Figure 3 Perturbation of gene expression in Tet1 paternal knockout embryos.
a, Heat-map representation of markedly downregulated genes (FC >1.5) in at least two E9.5 paternalKO embryos. Only those genes with significant change in at least two paternalKO embryos were shown. b, GO analysis of the 905 downregulated genes. c, Heat-map representation of markedly upregulated genes (FC >1.5) in at least E9.5 paternalKO embryos. d, GO analysis of the 635 upregulated genes.
Extended Data Figure 4 Hypermethylation in germline differentially methylated regions in Tet1 paternal knockout embryos and placentae, and Tet1 knockout sperm.
a, RT–qPCR analysis of Igf2r and Air in E9.5 embryos. The average value of control embryos (n = 13) is set as 1. Error bars indicate s.e.m. b, Bisulphite sequencing analysis of Air–Igf2r DMR of E9.5 embryos. The analysed region is indicated by a red line at the top of the diagram. Each CpG is represented by a circle with methylation and non-methylation represented by open and filled circles, respectively. The percentages of DNA methylation are indicated. c, qRT–PCR analysis of Impact in E9.5 embryos. The average value of control embryos (n = 13) is set as 1. Error bar indicates s.e.m. d, Bisulphite sequencing analysis of Impact DMR of E9.5 embryos. The analysed region is indicated by a red line at the top of the diagram. e, The weight of individual E19.5 paternalKO placentae (top) and qRT–PCR analysis of several imprinted gene expression patterns in each placenta. The average value of control placentae (n = 19) is set as 1. Asterisks indicate small placentae. Arrows indicate the samples showing a marked change in expression levels. Error bars indicate s.e.m. f, Bisulphite sequencing analysis of Peg3 DMR of E19.5 placentae. The analysed region is indicated at the top of the diagram. The percentages of DNA methylation are indicated. g, Bisulphite sequencing analysis of the Peg10 DMR in Tet1-heterozygous and knockout sperm. The analysed region is indicated by a red line at the top of the diagram.
Extended Data Figure 5 Summary of RRBS analysis.
a, b, Distribution of the level of methylation change for changed sites and promoters in E13.5 PGCs (a) and sperm (b) (cutoff, 10%). c, Cumulative distribution curve of methylation change in E13.5 PGCs for the sites showing methylation change in Tet1 knockout sperm. Note that methylation levels in E13.5 Tet1 knockout PGCs of both 422 hypermethylated sites in sperm (red) and 1,484 hypomethylated sites in sperm (blue) are higher than that of non-changed sites.
Extended Data Figure 6 Hypermethylation in Tet1 knockout E13.5 PGCs at imprinted genes and germline genes.
a, GO analysis of the genes with hypermethylated promoters in Tet1 knockout E13.5 PGCs. b, c, Representative DNA methylation profiles of selected imprinting genes (b) and germline genes (c) in E13.5 PGCs analysed by RRBS. Vertical grey lines represent the sequence read depth for each cytosine scored, and 30 reads were shown at most. The vertical red lines represent percentage of methylation for each cytosine scored, which range from 0% to 50%. For each gene, RefSeq exon organization (blue) and location of CGIs (green) are shown at the bottom.
Extended Data Figure 7 Hypermethylation in Tet1 knockout sperm at imprinted genes.
a, b, Mouse phenotype ontology enrichment of hypermethylated sites in Tet1 knockout sperm (a) and hypermethylated sites in both Tet1 knockout E13.5 PGCs and sperm (b). c, Representative DNA methylation profiles of selected imprinting genes in sperm analysed by RRBS. Vertical grey lines represent the sequence read depth for each cytosine scored, and 30 reads are shown at most. The vertical red lines represent percentage of methylation for each cytosine scored, which range from 0% to 50%. For each gene, RefSeq exon organization (blue) and location of CGIs (green) are shown at the bottom.
Extended Data Figure 8 Tet1 knockout affected loci are enriched in late demethylated regions in PGCs.
a, Methylation dynamics of the loci that are hypermethylated in Tet1 knockout E13.5 PGCs. Methylation levels were obtained from a previous study15. Middle lines in the coloured boxes indicate the median, box edges indicate the 25th/75th percentiles, and whiskers the 2.5th/97.5th percentiles. b, Percentages of hypermethylated promoters in Tet1 knockout PGCs among late demethylated promoters. The RRBS-covered promoters with >25% methylation in E9.5, E10.5 and E11.5 PGCs (ref. 14) were analysed. ***P < 1.0 × 10−15, **P < 0.01.
Extended Data Figure 9 Early embryonic lethality and dysregulation of imprinted gene expression in Tet1 paternal knockout mice generated by ref. 17 .
a, Representative image of paternalKO pups from the same litter showing big variation in body and placenta sizes. Embryos and their placentae were recovered by Caesarean section at the day of birth (E19.5). b, Average placental weights at E19.5. Error bars indicate s.e.m. **P < 0.01. c, Mating scores of Tet1+/− male × wild-type female (×Het) and Tet1−/− male × wild-type female (×KO) mice. Black dots indicate the number of pups for each litter recovered by Caesarean section at E19.5. Red lines indicate the mean litter size. d, Representative image of E10.5 Tet1 paternalKO embryos from the mating of wild-type female with Tet1 knockout male mice. The dotted oval indicates morphologically abnormal embryos. e, Left: E9.5 paternalKO embryos recovered from a single litter. Dotted oval indicates small embryos whose placentae are morphologically abnormal. Right: placentae of the corresponding embryos shown in the left panel. The red rectangle indicates morphologically abnormal placentae. f, Summary of embryonic abnormality. Asterisk indicates the numbers of morphologically abnormal embryos (E10.5) or placentae (E9.5). g, qRT–PCR analysis of selected imprinted genes in each of the E9.5 embryos from two litters (paternalKO 1 and paternalKO 2) are shown. Arrows indicate the samples showing marked change in expression level. The Ct values are normalized to the expression level of Gapdh.
Extended Data Figure 10 Defects in embryonic development and genomic imprinting in E19.5 Tet1 maternal knockout embryos and placentae.
a, Representative image of embryos and placentae from a single litter of E19.5 Tet1 maternalKO mice. Arrowheads indicate dead embryos whose placentae were morphologically normal. b, The ratio of dead embryos with relatively normal placentae. c, d, Box blot representation of the weight of embryos (c) and placentae (d). Middle lines in the coloured boxes indicate the medians, box edges indicate the 25th/75th percentiles, and whiskers the 0th/100th percentiles. e, qRT–PCR analysis of paternally imprinted genes in Tet1 maternalKO placentae. Arrows indicate the placentae whose embryos were found dead in utero, as indicated by arrowheads in a. The average value of control embryos (n = 19) is set as 1. Error bars indicate s.e.m. f, BS-seq revealed hypermethylation in Tet1 maternalKO mice in the IG-DMR locus. Note that methylation level in Tet1 maternalKO mice was significantly higher than in control mice, although IG-DMR gained methylation during placental development. Each dot represents the methylation level of each placenta. The numbers next to each dot indicate the number of clones sequenced. The average methylation levels are indicated by horizontal lines. The analysed region is indicated by a red line at the top of the diagram. g, BS-seq analysis of Rasgrf1 DMR of E19.5 placentae. The analysed region is indicated by a red line at the top of the diagram. Each CpG is represented by a circle with methylation and non-methylation represented by open and filled circles, respectively. The percentages of DNA methylation are indicated. *P < 0.05. **P < 0.01.
Extended Data Figure 11 Model explaining Tet1 paternal knockout phenotypes, using one maternal germline DMR as an example.
Black and white bars represent methylated and unmethylated state, respectively. In the control, the maternal allele is completely demethylated through Tet1-mediated imprinting erasure during PGC reprogramming. However, in Tet1 knockout PGCs, DMRs fail to be demethylated. The remaining methylation in the maternal allele is inherited to the next generation, which leads to imprinting defects.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and Supplementary References. (PDF 170 kb)
Supplementary Data
This file contains Supplementary Table 1. (XLS 22 kb)
Supplementary Data
This file contains Supplementary Table 2. (XLS 62 kb)
Supplementary Data
This file contains Supplementary Table 3. (XLS 661 kb)
Supplementary Data
This file contains Supplementary Table 4. (XLS 21 kb)
Supplementary Data
This file contains Supplementary Table 5. (XLS 2513 kb)
Supplementary Data
This file contains Supplementary Table 6. (XLS 729 kb)
Supplementary Data
This file contains Supplementary Table 7. (XLS 65 kb)
Supplementary Data
This file contains Supplementary Table 8. (XLS 37 kb)
Supplementary Data
This file contains Supplementary Table 9. (XLS 36 kb)
Rights and permissions
About this article
Cite this article
Yamaguchi, S., Shen, L., Liu, Y. et al. Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464 (2013). https://doi.org/10.1038/nature12805
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12805
This article is cited by
-
Epigenetic OCT4 regulatory network: stochastic analysis of cellular reprogramming
npj Systems Biology and Applications (2024)
-
The role of DNA hydroxymethylation and TET enzymes in placental development and pregnancy outcome
Clinical Epigenetics (2023)
-
Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development
Nature Genetics (2023)
-
Tet Enzyme-Mediated Response in Environmental Stress and Stress-Related Psychiatric Diseases
Molecular Neurobiology (2023)
-
Bend family proteins mark chromatin boundaries and synergistically promote early germ cell differentiation
Protein & Cell (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.