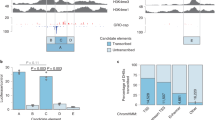
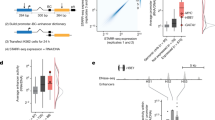
Abstract
Enhancers control the correct temporal and cell-type-specific activation of gene expression in multicellular eukaryotes. Knowing their properties, regulatory activity and targets is crucial to understand the regulation of differentiation and homeostasis. Here we use the FANTOM5 panel of samples, covering the majority of human tissues and cell types, to produce an atlas of active, in vivo-transcribed enhancers. We show that enhancers share properties with CpG-poor messenger RNA promoters but produce bidirectional, exosome-sensitive, relatively short unspliced RNAs, the generation of which is strongly related to enhancer activity. The atlas is used to compare regulatory programs between different cells at unprecedented depth, to identify disease-associated regulatory single nucleotide polymorphisms, and to classify cell-type-specific and ubiquitous enhancers. We further explore the utility of enhancer redundancy, which explains gene expression strength rather than expression patterns. The online FANTOM5 enhancer atlas represents a unique resource for studies on cell-type-specific enhancers and gene regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
DDBJ/GenBank/EMBL
Gene Expression Omnibus
Data deposits
The FANTOM5 atlas is accessible from http://fantom.gsc.riken.jp/5. FANTOM5 CAGE, RNA-seq and sRNA data have been deposited in DDBJ/EMBL/GenBank (accession codes DRA000991, DRA001101). Genome browser tracks for enhancers with user-definable expression specificity-constraints can be generated at http://enhancer.binf.ku.dk. Here, processed enhancer expression data, predefined enhancer tracks and motif finding results are also deposited. Blood-cell ChIP-seq data and CAGE data on exosome-depleted HeLa cells have been deposited in the NCBI GEO database (accession codes GSE40668, GSE49834).
References
Bulger, M. & Groudine, M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 339, 250–257 (2010)
Lenhard, B., Sandelin, A. & Carninci, P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nature Rev. Genet. 13, 233–245 (2012)
Banerji, J., Rusconi, S. & Schaffner, W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981)
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010)
Kodzius, R. et al. CAGE: cap analysis of gene expression. Nature Methods 3, 211–222 (2006)
The FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature http://dx.doi.org/10.1038/nature13182 (this issue).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Kheradpour, P. et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 23, 800–811 (2013)
Fort, A. et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nature Genet. (in the press)
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012)
Ntini, E. et al. Polyadenylation site–induced decay of upstream transcripts enforces promoter directionality. Nature Struct. Mol. Biol. 20, 923–928 (2013)
Almada, A. E., Wu, X., Kriz, A. J., Burge, C. B. & Sharp, P. A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013)
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012)
Kowalczyk, M. S. et al. Intragenic enhancers act as alternative promoters. Mol. Cell 45, 447–458 (2012)
Valen, E. et al. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nature Struct. Mol. Biol. 18, 1075–1082 (2011)
Taft, R. J. et al. Tiny RNAs associated with transcription start sites in animals. Nature Genet. 41, 572–578 (2009)
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008)
Rönnerblad, M. et al. Analysis of the DNA methylome and transcriptome in granulopoiesis reveal timed changes and dynamic enhancer methylation. Blood http://dx.doi.org/10.1182/blood-2013-02-482893 (in the press)
Biddie, S. C. et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 (2011)
Schmidt, D. et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20, 578–588 (2010)
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012)
Chepelev, I., Wei, G., Wangsa, D., Tang, Q. & Zhao, K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 22, 490–503 (2012)
Fraser, P., Pruzina, S., Antoniou, M. & Grosveld, F. Each hypersensitive site of the human beta-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev. 7, 106–113 (1993)
Dostie, J. et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006)
Barolo, S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 34, 135–141 (2012)
Schaffner, G., Schirm, S., Müller-Baden, B., Weber, F. & Schaffner, W. Redundancy of information in enhancers as a principle of mammalian transcription control. J. Mol. Biol. 201, 81–90 (1988)
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013)
Göring, H. H. H. et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nature Genet. 39, 1208–1216 (2007)
Hindorff, L. A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA 106, 9362–9367 (2009)
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012)
Maurano, M. T., Wang, H., Kutyavin, T. & Stamatoyannopoulos, J. A. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 8, e1002599 (2012)
Mercer, E. M. et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity 35, 413–425 (2011)
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013)
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011)
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010)
Gehrig, J. et al. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nature Methods 6, 911–916 (2009)
Kanamori-Katayama, M. et al. Unamplified cap analysis of gene expression on a single-molecule sequencer. Genome Res. 21, 1150–1159 (2011)
Khalil, A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11667–11672 (2009)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Hoffman, M. M. et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nature Methods 9, 473–476 (2012)
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nature Methods 9, 215–216 (2012)
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009)
Marshall, O. J. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20, 2471–2472 (2004)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 28, 511–515 (2010)
Preker, P. et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008)
Takahashi, H., Lassmann, T., Murata, M. & Carninci, P. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nature Protocols 7, 542–561 (2012)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Carninci, P. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genet. 38, 626–635 (2006)
Pham, T. H. et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood 119, e161–e171 (2012)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Schmidl, C. et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 19, 1165–1174 (2009)
Klug, M. & Rehli, M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 1, 127–130 (2006)
Rehli, M. et al. PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J. Biol. Chem. 275, 9773–9781 (2000)
Li, L. C. & Dahiya, R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 (2002)
Ehrich, M. et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl Acad. Sci. USA 102, 15785–15790 (2005)
Lin, J. Divergence measures based on the Shannon entropy. IEEE Trans. Inf. Theory 37, 145–151 (1991)
Hollander, M. & Wolfe, D. A. Nonparametric Statistical Methods (Wiley-Interscience, 1999)
Hothorn, T., Hornik, K., Van De Wiel, M. A. & Zeileis, A. A Lego system for conditional inference. Am. Stat. 60, 257–263 (2006)
Buckner, J. et al. The gputools package enables GPU computing in R. Bioinformatics 26, 134–135 (2010)
Ellingsen, S. et al. Large-scale enhancer detection in the zebrafish genome. Development 132, 3799–3811 (2005)
Meng, A., Tang, H., Ong, B. A., Farrell, M. J. & Lin, S. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc. Natl Acad. Sci. USA 94, 6267–6272 (1997)
Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). (Univ. Oregon Press, 1995)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2008)
Zuber, V. & Strimmer, K. High-dimensional regression and variable selection using CAR scores. Stat. Appl. Genet. Mol. Biol. 10, 1–27 (2011)
Chevan, A. & Sutherland, M. Hierarchical partitioning. Am. Stat. 45, 90–96 (1991)
Groemping, U. Relative importance for linear regression in R: the package relaimpo. J. Stat. Softw. 17, 1–27 (2006)
Johnson, A. D. et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24, 2938–2939 (2008)
The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010)
Rhead, B. et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38, D613–D619 (2010)
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004)
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010)
Acknowledgements
FANTOM5 was made possible by a Research Grant for RIKEN Omics Science Center from MEXT to Y.H. and a Grant of the Innovative Cell Biology by Innovative Technology (Cell Innovation Program) from the MEXT, Japan, to Y.H. The A.S. group was supported by funds from the European Research Council FP7/2007-2013/ERC no. 204135, the Novo Nordisk and Lundbeck foundations. Work in the M.R. group was funded by grants from the Deutsche Forschungsgemeinschaft (RE 1310/7, 11, 13) and Rudolf Bartling Stiftung. F.M. and I.M.E. were supported by “BOLD” Marie Curie ITN and “ZF- Health” Integrated project of the European Commission. We thank S. Noma, M. Sakai and H. Tarui for RNA-seq and sRNA-seq preparation, RIKEN GeNAS for generation and sequencing of the Heliscope CAGE libraries, Illumina RNA-seq and sRNA-seq, the Copenhagen National High-throughput DNA Sequencing Center for Illumina CAGE-seq, M. Edinger, P. Hoffmann and R. Eder for cell sorting, A. Albrechtsen, I. Moltke, W. Wasserman for advice, and the Netherlands Brain Bank for post-mortem human brain material.
Author information
Authors and Affiliations
Consortia
Contributions
R.A., I.H., E.A., E.V., K.L., Y.C., B.L., X.Z., M.J., H.K., T.F.M., T.L., N.B., O.R., A.M.B. , J.K.B, C.J.M, N.R., F.O.B., M.R., A.S. made the computational analysis. J.B., M.B., T.L., H.K., N.K., J.K., H.S., M.I., C.O.D, A.R.R.F., P.C., Y.H. prepared and pre-processed CAGE and/or RNA-seq libraries. E.N., P.R.A., T.H.J., J.B., M.B. made the knockdown experiments followed by CAGE. C.G., C.S., L.S., J.R., D.G., M.R. made the blood cell ChIP experiments, methylation assays and in vitro blood cell validations. T.S., C.G., Y.I., Y.S., E.F., S.M., Y.N., A.R.R.F., P.C. and H.S. made the HeLa/HepG2 in vitro validations. I.M.E., R.A., A.S., F.M. designed and carried out zebrafish in vivo tests. R.A., C.G., I.H., C.S., E.A., E.V., F.M., I.M.E., P.C., A.R.R.F, M.B., J.B., A.L., C.D., D.A.H., P.H., M.R., A.S. interpreted results. R.A., C.G., I.H., E.V., I.M.E., J.B., F.M., D.A.H., M.R., A.S. wrote the paper with input from all authors. M.R. and A.S. coordinated and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of authors and affiliations appears in the Supplementary Information.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary Table Legends, Supplementary References, a full list of members of the FANTOM consortium and Supplementary Figures 1-33. (PDF 19934 kb)
This file contains Supplementary Text, Supplementary Table Legends, Supplementary References, a full list of members of the FANTOM consortium and Supplementary Figures 1-33.
This file contains Supplementary Tables 1-16 - see Supplementary Information document for legends. (ZIP 436 kb)
Rights and permissions
About this article
Cite this article
Andersson, R., Gebhard, C., Miguel-Escalada, I. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). https://doi.org/10.1038/nature12787
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12787
This article is cited by
-
Tissue-specific RNA Polymerase II promoter-proximal pause release and burst kinetics in a Drosophila embryonic patterning network
Genome Biology (2024)
-
Boundary stacking interactions enable cross-TAD enhancer–promoter communication during limb development
Nature Genetics (2024)
-
Selective gene expression maintains human tRNA anticodon pools during differentiation
Nature Cell Biology (2024)
-
Biological basis of extensive pleiotropy between blood traits and cancer risk
Genome Medicine (2024)
-
An overview of artificial intelligence in the field of genomics
Discover Artificial Intelligence (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.