Abstract
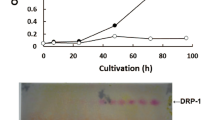
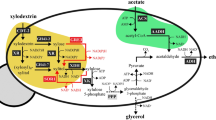
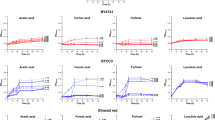
The increasing demands placed on natural resources for fuel and food production require that we explore the use of efficient, sustainable feedstocks such as brown macroalgae. The full potential of brown macroalgae as feedstocks for commercial-scale fuel ethanol production, however, requires extensive re-engineering of the alginate and mannitol catabolic pathways1,2,3 in the standard industrial microbe Saccharomyces cerevisiae. Here we present the discovery of an alginate monomer (4-deoxy-l-erythro-5-hexoseulose uronate, or DEHU) transporter from the alginolytic eukaryote Asteromyces cruciatus4. The genomic integration and overexpression of the gene encoding this transporter, together with the necessary bacterial alginate and deregulated native mannitol catabolism genes, conferred the ability of an S. cerevisiae strain to efficiently metabolize DEHU and mannitol. When this platform was further adapted to grow on mannitol and DEHU under anaerobic conditions, it was capable of ethanol fermentation from mannitol and DEHU, achieving titres of 4.6% (v/v) (36.2 g l−1) and yields up to 83% of the maximum theoretical yield from consumed sugars. These results show that all major sugars in brown macroalgae can be used as feedstocks for biofuels and value-added renewable chemicals in a manner that is comparable to traditional arable-land-based feedstocks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
08 January 2014
A present address was added for author Avinash Gill.
References
Wargacki, A. J. et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335, 308–313 (2012)
Santos, C. N., Regitsky, D. D. & Yoshikuni, Y. Implementation of stable and complex biological systems through recombinase-assisted genome engineering. Nature Comm. 4, 2503 (2013)
Roesijadi, G., Jones, S. B., Snowden-Swan, L. J. & Zhu, Y. PNNL-19944: Macroalgae as a Biomass Feedstock: A Preliminary Analysis (Pacific Northwest National Laboratory, 2010)
Schaumann, K. & Weide, G. Efficiency of uronic acid uptake in marine alginate-degrading fungi. Helgol. Meersunters. 49, 159–167 (1995)
World Population Prospects. The 2012 Revision, Volume 1 Comprehensive Table (United Nations, 2013)
International Energy Agency. World Energy Outlook 2012. (2012)
Interagency Agricultural Projections Committee. USDA Agricultural Projections to 2021. (2012)
Valdes, C. Can Brazil meet the world’s growing need for ethanol? AMBER WAVES 9, 38–45 (2011)
Renewable Fuels Association. http://www.ethanolrfa.org/pages/statistics#F (2013)
Food and Agriculture Organization of United Nations. How to Feed the World in 2050. (2009)
United Nations Educational, Scientific and Cultural Organization. The United Nations World Water Development Report 4: Managing Water under Uncertainty and Risk. (2012)
Stewart, W. M., Dibbb, D. W., Johnston, A. E. & Smyth, T. J. The contribution of commercial fertilizer nutrients to food production. Agron. J. 97, 1–6 (2005)
Chapman, V. J. Seaweeds and Their Uses, 2nd edn (The Camelot Press, 1970)
Horn, S. J., Aasen, M. & Østgaard, K. Ethanol production from seaweed extract. J. Ind. Microbiol. Biotechnol. 25, 249–254 (2000)
Nakamura, C. E. & Whited, G. M. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14, 454–459 (2003)
Yim, H. et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nature Chem. Biol. 7, 445–452 (2011)
Matrin Patel, M. C. et al. Medium and long-term opportunities and risks of the biotechnological production of bulk chemicals from renewable resources - the potential of white biotechnology. (The Brew Project, 2006)
Quain, D. E. & Boulton, C. A. Growth and metabolism of mannitol by strains of Saccharomyces cerevisiae. J. Gen. Microbiol. 133, 1675–1684 (1987)
Noiraud, N., Maurousset, L. & Lemoine, R. Identification of a mannitol transporter, AgMaT1, in celery phloem. Plant Cell 13, 695–705 (2001)
Mairta, P. K. & Lobo, Z. A kinetic study of glycolytic enzyme synthesis in yeast. J. Biol. Chem. 246, 425–468 (1971)
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotechnol. 29, 644–652 (2011)
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578 (2012)
Hilditch, S. Identification of the Fungal Catabolic d -galacturonate Pathway. PhD thesis, Univ. Helsinki. (2010)
Wheeler, K. A., Lamb, H. K. & Hawkins, A. R. Control of metabolic flux through the quinate pathway in Aspergillus nidulans. Biochem. J. 315, 195–205 (1996)
Preiss, J. & Ashwell, G. Alginic acid metabolism in bacteria. II. The enzymatic reduction of 4-deoxy-l-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-d-gluconic acid. J. Biol. Chem. 237, 317–321 (1962)
Migneault, I., Dartiguenave, C., Bertrand, M. J. & Waldron, K. C. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 37, 790–796, 798–802 (2004)
Jongkees, S. A. & Withers, S. G. Glycoside cleavage by a new mechanism in unsaturated glucuronyl hydrolases. J. Am. Chem. Soc. 133, 19334–19337 (2011)
Lau, M. W. & Dale, B. E. Cellulosic ethanol production from AFEX-treated corn stover using Saccharomyces cerevisiae 424A(LNH-ST). Proc. Natl Acad. Sci. USA 106, 1368–1373 (2009)
Peralta-Yahya, P. P., Zhang, F., del Cardayre, S. B. & Keasling, J. D. Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012)
Lee, J. W. et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nature Chem. Biol. 8, 536–546 (2012)
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008)
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnol. 31, 46–53 (2013)
Acknowledgements
We thank R. Schekman, J. H. D. Cate and P. A. Silver for critical discussion and suggestions. This work is supported by the DOE under an Advanced Research Projects Agency–Energy (ARPA-E) award (DE-AR0000006), by CORFO INNOVA CHILE (código 09CTEI-6866), and by Statoil ASA. This report was prepared as an account of work sponsored by an agency of the US government. Neither the US government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favouring by the US government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the US government or any agency thereof.
Author information
Authors and Affiliations
Contributions
Y.Y. conceived the overall project. C.S., Y.Y., M.E.-N., N.L.O., R.B.B., H.K.K. and V.R. supervised the overall project. R.M.R., S.R.C., P.D.D., Y.M. and A.G. characterised the A. cruciatus alginate degradation profiles. A.M.E.F., M.E.-N., P.K., L.L., M.Y.C. and S.A.T. constructed all S. cerevisiae host strains for screening. R.M.R. and P.D.D. prepared A. cruciatus genomic DNA and total RNA for the high-throughput sequencing analysis. D.D.R., R.M.R. and Y.Y. assembled and analysed the RNA-seq data. C.N.S.S., P.T., M.E.-N., J.G. and C.J.P. constructed and screened the A. cruciatus cDNA library. M.E.-N. and J.L.V. identified yeast strains that can grow on mannitol and performed microarray analyses to isolate mannitol catabolism genes. A.M.E.F., M.E.-N., L.L., M.Y.C., P.D.D. and J.G. constructed all S. cerevisiae host strains for fermentation experiments. Y.Y., D.D.B., M.E.-N. and A.C. carried out all adaptation and ethanol fermentation experiments. A.H. and P.S. developed and carried out biochemical assays for enzyme characterisation. S.R.C., P.S., Y.Y., D.D.B., B.L. and A.H. performed all necessary analytical experiments and analysed all fermentation samples. L.P., C.L., E.J.K., N.L.O. and D.J.M. conducted sample preparation and the NMR analysis of the DEHU structure. C.L., P.S., D.D.B. and Y.Y. prepared media for fermentation, Y.Y., M.E.-N., A.M.E.F., C.S., D.D.B., C.N.S.S. and L.P. wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Y.Y., J.L.V., S.R.C., V.R., S.A.T., M.E.-N., R.M.R., A.G., A.H., C.N.S.S., D.D.R, C.S., R.B.B. and A.M.E.F. are co-inventors of a patent application entitled “Methods and Compositions for Growth of Yeast on Alginate” (PCT/US2013/021328).
Supplementary information
Supplementary Information
This file contains a list of abbreviations, Supplementary Discussion, Supplementary Tables 1-12, Supplementary Figures 1-15, Supplementary Sequence and Supplementary References. (PDF 2151 kb)
Rights and permissions
About this article
Cite this article
Enquist-Newman, M., Faust, A., Bravo, D. et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 505, 239–243 (2014). https://doi.org/10.1038/nature12771
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12771
This article is cited by
-
Component analysis and utilization strategy of brown macroalgae as promising feedstock for sugar platform-based marine biorefinery
Biotechnology and Bioprocess Engineering (2024)
-
Characterization and Mechanism Study of a Novel PL7 Family Exolytic Alginate Lyase from Marine Bacteria Vibrio sp. W13
Applied Biochemistry and Biotechnology (2024)
-
Deciphering of the Mannitol Metabolism Pathway in Clostridium tyrobutyricum ATCC 25755 by Comparative Transcriptome Analysis
Applied Biochemistry and Biotechnology (2023)
-
A First Marine Vibrio Biocatalyst to Produce Ethanol from Alginate, which is a Rich Polysaccharide in Brown Macroalgal Biomass
Current Microbiology (2023)
-
Diversity of culturable alginate lyase-excreting bacteria associated with Sargassum
Acta Oceanologica Sinica (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.