Abstract
CAAX proteins have essential roles in multiple signalling pathways, controlling processes such as proliferation, differentiation and carcinogenesis1. The ∼120 mammalian CAAX proteins function at cellular membranes and include the Ras superfamily of small GTPases, nuclear lamins, the γ-subunit of heterotrimeric GTPases, and several protein kinases and phosphatases2. The proper localization of CAAX proteins to cell membranes is orchestrated by a series of post-translational modifications of the carboxy-terminal CAAX motifs3 (where C is cysteine, A is an aliphatic amino acid and X is any amino acid). These reactions involve prenylation of the cysteine residue, cleavage at the AAX tripeptide and methylation of the carboxyl-prenylated cysteine residue. The major CAAX protease activity is mediated by Rce1 (Ras and a-factor converting enzyme 1), an intramembrane protease (IMP) of the endoplasmic reticulum4,5. Information on the architecture and proteolytic mechanism of Rce1 has been lacking. Here we report the crystal structure of a Methanococcus maripaludis homologue of Rce1, whose endopeptidase specificity for farnesylated peptides mimics that of eukaryotic Rce1. Its structure, comprising eight transmembrane α-helices, and catalytic site are distinct from those of other IMPs. The catalytic residues are located ∼10 Å into the membrane and are exposed to the cytoplasm and membrane through a conical cavity that accommodates the prenylated CAAX substrate. We propose that the farnesyl lipid binds to a site at the opening of two transmembrane α-helices, which results in the scissile bond being positioned adjacent to a glutamate-activated nucleophilic water molecule. This study suggests that Rce1 is the founding member of a novel IMP family, the glutamate IMPs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Winter-Vann, A. M. & Casey, P. J. Post-prenylation-processing enzymes as new targets in oncogenesis. Nature Rev. Cancer 5, 405–412 (2005)
Prior, I. A. & Hancock, J. F. Ras trafficking, localization and compartmentalized signalling. Semin. Cell Dev. Biol. 23, 145–153 (2012)
Ahearn, I. M., Haigis, K., Bar-Sagi, D. & Philips, M. R. Regulating the regulator: post-translational modification of RAS. Nature Rev. Mol. Cell Biol. 13, 39–51 (2012)
Boyartchuk, V. L., Ashby, M. N. & Rine, J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275, 1796–1800 (1997)
Schmidt, W. K., Tam, A., Fujimura-Kamada, K. & Michaelis, S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl Acad. Sci. USA 95, 11175–11180 (1998)
Michaelis, S. & Barrowman, J. Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol. Mol. Biol. Rev. 76, 626–651 (2012)
Wang, Y., Zhang, Y. & Ha, Y. Crystal structure of a rhomboid family intramembrane protease. Nature 444, 179–180 (2006)
Feng, L. et al. Structure of a site-2 protease family intramembrane metalloprotease. Science 318, 1608–1612 (2007)
Li, X. et al. Structure of a presenilin family intramembrane aspartate protease. Nature 493, 56–61 (2013)
Pryor, E. E., Jr et al. Structure of the integral membrane protein CAAX protease Ste24p. Science 339, 1600–1604 (2013)
Quigley, A. et al. The structural basis of ZMPSTE24-dependent laminopathies. Science 339, 1604–1607 (2013)
Hu, J., Xue, Y., Lee, S. & Ha, Y. The crystal structure of GXGD membrane protease FlaK. Nature 475, 528–531 (2011)
Pei, J. & Grishin, N. V. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26, 275–277 (2001)
Kjos, M., Snipen, L., Salehian, Z., Nes, I. F. & Diep, D. B. The Abi proteins and their involvement in bacteriocin self-immunity. J. Bacteriol. 192, 2068–2076 (2010)
Dolence, J. M., Steward, L. E., Dolence, E. K., Wong, D. H. & Poulter, C. D. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry 39, 4096–4104 (2000)
Plummer, L. J. et al. Mutational analysis of the Ras converting enzyme reveals a requirement for glutamate and histidine residues. J. Biol. Chem. 281, 4596–4605 (2006)
Bergo, M. O. et al. Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol. Cell. Biol. 22, 171–181 (2002)
Wahlstrom, A. M. et al. Rce1 deficiency accelerates the development of K-RAS-induced myeloproliferative disease. Blood 109, 763–768 (2007)
Bergo, M. O. et al. On the physiological importance of endoproteolysis of CAAX proteins: heart-specific RCE1 knockout mice develop a lethal cardiomyopathy. J. Biol. Chem. 279, 4729–4736 (2004)
Christiansen, J. R., Kolandaivelu, S., Bergo, M. O. & Ramamurthy, V. RAS-converting enzyme 1-mediated endoproteolysis is required for trafficking of rod phosphodiesterase 6 to photoreceptor outer segments. Proc. Natl Acad. Sci. USA 108, 8862–8866 (2011)
Otto, J. C., Kim, E., Young, S. G. & Casey, P. J. Cloning and characterization of a mammalian prenyl protein-specific protease. J. Biol. Chem. 274, 8379–8382 (1999)
Hollander, I., Frommer, E. & Mallon, R. Human Ras-converting enzyme (hRCE1) endoproteolytic activity on K-Ras-derived peptides. Anal. Biochem. 286, 129–137 (2000)
Hollander, I. J., Frommer, E., Aulabaugh, A. & Mallon, R. Human Ras converting enzyme endoproteolytic specificity at the P2′ and P3′ positions of K-Ras-derived peptides. Biochim. Biophys. Acta 1649, 24–29 (2003)
Wu, Z. et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nature Struct. Mol. Biol. 13, 1084–1091 (2006)
Baker, R. P., Young, K., Feng, L., Shi, Y. & Urban, S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc. Natl Acad. Sci. USA 104, 8257–8262 (2007)
Ben-Shem, A., Fass, D. & Bibi, E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl Acad. Sci. USA 104, 462–466 (2007)
Wang, Y. & Ha, Y. Open-cap conformation of intramembrane protease GlpG. Proc. Natl Acad. Sci. USA 104, 2098–2102 (2007)
Fujinaga, M., Cherney, M. M., Oyama, H., Oda, K. & James, M. N. The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum. Proc. Natl Acad. Sci. USA 101, 3364–3369 (2004)
Vinothkumar, K. R. et al. The structural basis for catalysis and substrate specificity of a rhomboid protease. EMBO J. 29, 3797–3809 (2010)
Trueblood, C. E. et al. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol. Cell. Biol. 20, 4381–4392 (2000)
Day, P. W. et al. A monoclonal antibody for G protein-coupled receptor crystallography. Nature Methods 4, 927–929 (2007)
Hino, T. et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature 482, 237–240 (2012)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. D 66, 470–478 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Raveh, B., London, N., Zimmerman, L. & Schueler-Furman, O. Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS ONE 6, e18934 (2011)
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005)
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Eswar, N. et al. in Current Protocols in Bioinformatics Ch. 5 (Wiley, 2006)
Provencher, S. W. & Glockner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 (1981)
Sreerama, N. & Woody, R. W. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209, 32–44 (1993)
Sreerama, N., Venyaminov, S. Y. & Woody, R. W. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8, 370–380 (1999)
Whitmore, L. & Wallace, B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 (2004)
Lees, J. G., Miles, A. J., Wien, F. & Wallace, B. A. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics 22, 1955–1962 (2006)
Alexandrov, A. I., Mileni, M., Chien, E. Y., Hanson, M. A. & Stevens, R. C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008)
Riou, P. et al. 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell 153, 640–653 (2013)
Acknowledgements
This work was funded by a Cancer Research UK grant to D.B. Part of this work was supported by the research acceleration program of the Japan Science and Technology agency and by BBSRC BB/G023425/1 (S.I.). We thank staff at the I04-1 Diamond Light Source for help with data collection, J. Yang for advice and discussions, I. De Moraes for support and T. Daviter for help with the circular dichroism experiments.
Author information
Authors and Affiliations
Contributions
R.B.D. and I.M. cloned and established the purification protocols for MmRce1. I.M. purified MmRce1 and prepared MmRce1–Fab co-crystals. S.O. and S.I. developed MmRce1-specific monoclonal antibodies. I.M., K.K., N.C. and D.B. collected crystallographic data. K.K. and I.M. determined the MmRce1 structure. I.M. and Z.Z. generated MmRce1 mutants. I.M. performed the kinetic and stability assays. K.K. performed the protein modelling and peptide docking. A.J.T. and S.J.H. performed the mass spectrometry analysis. N.O′R. and G.B. synthesized geranylgernanylated peptides. I.M. and D.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
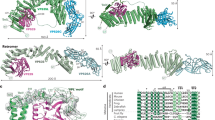
Extended Data Figure 1 Structure of the MmRce1–Fab645-2 complex.
Overall ribbon diagram of the MmRce1–Fab645-2 complex, with MmRce1 viewed parallel to the membrane. MmRce1 is depicted in blue, the antibody Fab fragment heavy chain (Fab(H)) in yellow, and the antibody Fab fragment light chain (Fab(L)) in green. MmRce1 interacts with Fab645-2 through its cytoplasmic side.
Extended Data Figure 2 Fluorescence resonance energy transfer (FRET) proteolytic assay of purified MmRce1.
The FRET assay was performed using a CAAX synthetic peptide modelled on the C terminus of RhoA (Fig. 1b and Methods). The plotted graph was fitted to a nonlinear regression one-phase decay equation, because hydrolysis of the peptide by MmRce1 obeyed Michaelis–Menten kinetics. The apparent binding constant (Km) was 19.7 ± 1.0 μM, and the apparent turnover constant (kcat) was 0.175 ± 0.0027 s−1. The data are presented as the mean + s.d. of n = 3 experiments. The fluorescence conversion factor (relative fluorescence units versus peptide concentration; inset curve) was determined by measuring the FRET between DABCYL and EDANS at different peptide concentrations (10–90 μM) and plotting the standard curve (fluorescence versus concentration). The data are presented as the mean + s.d. of n = 3 experiments, and R2 indicates the quality of the fit.
Extended Data Figure 3 Structure-based multiple sequence alignment.
Structure-based multiple sequence alignment of Rce1 homologues representing all three domains of life: Homo sapiens RCE1 (HsRce1; UniProt ID Q9Y256), Saccharomyces cerevisiae Rce1p (ScRce1; UniProt ID Q03530), Streptomyces coelicolor Rce1 (ScoRce1; UniProt ID Q9XAK4), Methanococcus maripaludis Rce1 (MmRce1; UniProt ID Q6LZY8) and Lactobacillus plantarum Rce1 (LpRce1; UniProt ID C6VK86). The transmembrane helices are depicted as cyan barrels. The residues that were mutated and disrupt activity are indicated by green and orange arrows: the green arrows indicate catalytic residues, and the orange arrow indicates a putative farnesyl lipid-binding residue in TM4. HsRce1 and MmRce1 have 15% sequence identity.
Extended Data Figure 4 Effects of zinc and chelating factors on the activity and stability of MmRce1.
Comparison of the proteolytic activities of wild-type MmRce1 and MmRce1 incubated with a molar excess of ZnSO4, EDTA, 1,10-phenanthroline or 1,7-phenanthroline. 1,7-Phenanthroline, a stereoisomer of 1,10-phenanthroline that does not chelate Zn2+, inactivated MmRce1 to the same extent as 1,10-phenanthroline, which does chelate Zn2+. A molar excess of Zn2+ (5 mM) did not reverse the inactivating effect of 0.3 mM 1,10-phenanthroline on MmRce1. The data are presented as the mean + s.d. of n = 3 experiments. b, c, Representative derivative melt profiles of MmRce1 incubated without (black) and with 5 mM 1,7-phenanthroline (red) or 1,10-phenanthroline (blue) (b). Experiments were performed over a temperature range starting from 10 °C and ending at 95 °C. The melting temperatures (Tm) were taken from the minimum derivative plot (c), plotted as the negative of the first derivative of the normal fluorescent curve, converted by Protein Thermal Shift software. The Tm values were determined as the mean + s.d. of n = 3 replicates. d, e, Two views showing ribbon and surface representations of MmRce1 (molecule C from the asymmetric unit). The side chain of C213 is shown as an orange stick model. C213 is buried within the structure and inaccessible unless the protein unfolds and/or undergoes transient ‘breathing’ motions.
Extended Data Figure 5 Thermal stability comparisons.
a, b, Representative melting curves of wild-type MmRce1 and mutants obtained from the CPM-based thermostability assay (Methods) fitted to a Boltzmann sigmoidal equation. The raw data for each protein are shown as dots, and the fitted curves are shown as black lines. The measurements were obtained over a period of 120 min at 40 °C. c–f, Representative derivative melt profiles of wild-type MmRce1 and mutants (c) and wild-type MmRce1 and MmRce1 incubated with increasing amounts of urea (1–5 M) (e). Experiments were performed over a temperature range starting from 10 °C and ending at 95 °C. The melting temperatures (Tm) were taken from the minimum derivative plot (d, f), plotted as the negative of the first derivative of the normal fluorescent curve, converted by Protein Thermal Shift software. The Tm values were determined as the mean + s.d. of n = 3 replicates.
Extended Data Figure 6 Circular dichroism.
a, Far ultraviolet circular dichroism spectra of wild-type MmRce1 and mutants (E140A, H173A, H227A, N231A, L45W and L132W). The measurements were made at 25 °C in 20 mM MES (pH 6.5), 100 mM KF, 0.029% (w/v) UDM and 0.86% (w/v) OG. Contributions to the spectra by the buffer were subtracted using control scans. All curves represent Δε measured in units of l mol−1 cm−1. b, Estimate of the proportion of α-helical structure in wild-type MmRce1 and mutants. Analysis of the circular dichroism spectra was performed using CONTIN/LL. The error bars represent a presumed inaccuracy in the protein concentration determination of 10%.
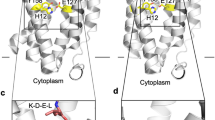
Extended Data Figure 7 The proposed catalytic mechanism of MmRce1.
The catalytic mechanism and overall structure of MmRce1 compared with those of GlpG (PDB ID 2O7L)27, PSH (presenilin/SPP homologue) (PDB ID 4HYC)9 and ZMPSTE24 (PDB ID 4AW6)11. The MmRce1 water molecule, which is hydrogen-bonded to both E140 and H173, is the nucleophile. The general base is the carboxylate of E140. H227 and N231 (both marked “oxy”) are proposed to stabilize the oxyanion transition state. Dashed lines represent hydrogen bonds or other electrostatic interactions. The arrows indicate the movement of electron pairs. The ribbon diagrams of MmRce1, GlpG, PSH and ZMPSTE24 are viewed parallel to the membrane, oriented from the top (the endoplasmic reticulum (ER) lumen) to the bottom (cytoplasm).
Extended Data Figure 8 Disruption of the farnesyl lipid-binding site.
a, b, Both L-to-W mutants are modelled, and the mutations are depicted in orange as side-chain stick models and surface representations. c, The proteolytic activity of wild-type MmRce1 and mutants towards a farnesylated peptide. The data are presented as the mean + s.d. of n = 3 experiments. d, Superposition of the MmRce1 crystal structure (orange) and a human RCE1 (HsRce1) homology model (blue). e, Cartoon representation of the HsRce1 homology model with a geranylgeranylated peptide modelled at its catalytic site. The geranylgeranylated peptide (GAKASGC(geranylgeranyl)LVS) is depicted as a green ribbon with the N and C termini labelled, and the geranylgeranyl lipid is depicted as a yellow ball-and-stick model.
Extended Data Figure 9 Final 2Fo−Fc electron density maps.
a, Stereoview showing 2Fo−Fc electron density calculated after the final refinement run (contoured at the 1σ level) (grey), with the final MmRce1 model overlaid (blue). b, Stereoview of the 2Fo−Fc electron density (contoured at the 1σ level) at the MmRce1 catalytic site. The catalytic residues (E140 and H173), the catalytic water molecule (W) and the oxyanion residues (H227 and N231) are labelled, as well as a second water molecule (W2) that interacts directly with W. c, Stereoview of 2Fo−Fc electron density in the vicinity of the MmRce1 transmembrane helices.
Rights and permissions
About this article
Cite this article
Manolaridis, I., Kulkarni, K., Dodd, R. et al. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature 504, 301–305 (2013). https://doi.org/10.1038/nature12754
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12754
This article is cited by
-
The generation of detergent-insoluble clipped fragments from an ERAD substrate in mammalian cells
Scientific Reports (2023)
-
Helical stability of the GnTV transmembrane domain impacts on SPPL3 dependent cleavage
Scientific Reports (2022)
-
Bioinformatic mapping of a more precise Aspergillus niger degradome
Scientific Reports (2021)
-
Post-translational modification of KRAS: potential targets for cancer therapy
Acta Pharmacologica Sinica (2021)
-
Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization
Nature Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.