Abstract
Glucose homeostasis is a vital and complex process, and its disruption can cause hyperglycaemia and type II diabetes mellitus1. Glucokinase (GK), a key enzyme that regulates glucose homeostasis, converts glucose to glucose-6-phosphate2,3 in pancreatic β-cells, liver hepatocytes, specific hypothalamic neurons, and gut enterocytes4. In hepatocytes, GK regulates glucose uptake and glycogen synthesis, suppresses glucose production3,5, and is subject to the endogenous inhibitor GK regulatory protein (GKRP)6,7,8. During fasting, GKRP binds, inactivates and sequesters GK in the nucleus, which removes GK from the gluconeogenic process and prevents a futile cycle of glucose phosphorylation. Compounds that directly hyperactivate GK (GK activators) lower blood glucose levels and are being evaluated clinically as potential therapeutics for the treatment of type II diabetes mellitus1,9,10. However, initial reports indicate that an increased risk of hypoglycaemia is associated with some GK activators11. To mitigate the risk of hypoglycaemia, we sought to increase GK activity by blocking GKRP. Here we describe the identification of two potent small-molecule GK–GKRP disruptors (AMG-1694 and AMG-3969) that normalized blood glucose levels in several rodent models of diabetes. These compounds potently reversed the inhibitory effect of GKRP on GK activity and promoted GK translocation both in vitro (isolated hepatocytes) and in vivo (liver). A co-crystal structure of full-length human GKRP in complex with AMG-1694 revealed a previously unknown binding pocket in GKRP distinct from that of the phosphofructose-binding site. Furthermore, with AMG-1694 and AMG-3969 (but not GK activators), blood glucose lowering was restricted to diabetic and not normoglycaemic animals. These findings exploit a new cellular mechanism for lowering blood glucose levels with reduced potential for hypoglycaemic risk in patients with type II diabetes mellitus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Verspohl, E. J. Novel pharmacological approaches to the treatment of type 2 diabetes. Pharmacol. Rev. 64, 188–237 (2012)
Agius, L. Glucokinase and molecular aspects of liver glycogen metabolism. Biochem. J. 414, 1–18 (2008)
Matschinsky, F. M. Glucokinase as glucose sensor and metabolic signal generator in pancreatic beta-cells and hepatocytes. Diabetes 39, 647–652 (1990)
Jetton, T. L. et al. Analysis of upstream glucokinase promoter activity in transgenic mice and identification of glucokinase in rare neuroendocrine cells in the brain and gut. J. Biol. Chem. 269, 3641–3654 (1994)
Iynedjian, P. B. Mammalian glucokinase and its gene. Biochem. J. 293, 1–13 (1993)
Van Schaftingen, E., Vandercammen, A., Detheux, M. & Davies, D. R. The regulatory protein of liver glucokinase. Adv. Enzyme Regul. 32, 133–148 (1992)
Vandercammen, A. & Van Schaftingen, E. The mechanism by which rat liver glucokinase is inhibited by the regulatory protein. Eur. J. Biochem. 191, 483–489 (1990)
Anderka, O. et al. Biophysical characterization of the interaction between hepatic glucokinase and its regulatory protein: impact of physiological and pharmacological effectors. J. Biol. Chem. 283, 31333–31340 (2008)
Coghlan, M. & Leighton, B. Glucokinase activators in diabetes management. Expert Opin. Investig. Drugs 17, 145–167 (2008)
Grimsby, J., Berthel, S. J. & Sarabu, R. Glucokinase activators for the potential treatment of type 2 diabetes. Curr. Top. Med. Chem. 8, 1524–1532 (2008)
Meininger, G. E. et al. Effects of MK-0941, a novel glucokinase activator, on glycemic control in insulin-treated patients with type 2 diabetes. Diabetes Care 34, 2560–2566 (2011)
Grimsby, J. et al. Allosteric activators of glucokinase: potential role in diabetes therapy. Science 301, 370–373 (2003)
Detheux, M., Vandercammen, A. & Van Schaftingen, E. Effectors of the regulatory protein acting on liver glucokinase: a kinetic investigation. Eur. J. Biochem. 200, 553–561 (1991)
Veiga-da-Cunha, M., Sokolova, T., Opperdoes, F. & Van Schaftingen, E. Evolution of vertebrate glucokinase regulatory protein from a bacterial N-acetylmuramate 6-phosphate etherase. Biochem. J. 423, 323–332 (2009)
Veiga-da-Cunha, M. & Van Schaftingen, E. Identification of fructose 6-phosphate- and fructose 1-phosphate-binding residues in the regulatory protein of glucokinase. J. Biol. Chem. 277, 8466–8473 (2002)
Fyfe, M. C. et al. Glucokinase activator PSN-GK1 displays enhanced antihyperglycaemic and insulinotropic actions. Diabetologia 50, 1277–1287 (2007)
Agius, L. & Peak, M. Intracellular binding of glucokinase in hepatocytes and translocation by glucose, fructose and insulin. Biochem. J. 296, 785–796 (1993)
Diabetes Genetics Initiative of Broad Institute of Harvard and MIT et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007)
Eiki, J. et al. Pharmacokinetic and pharmacodynamic properties of the glucokinase activator MK-0941 in rodent models of type 2 diabetes and healthy dogs. Mol. Pharmacol. 80, 1156–1165 (2011)
Grimsby, J. et al. Characterization of glucokinase regulatory protein-deficient mice. J. Biol. Chem. 275, 7826–7831 (2000)
Pfefferkorn, J. A. et al. Discovery of (S)-6-(3-cyclopentyl-2-(4-(trifluoromethyl)-1H-imidazol-1-yl)propanamido)nicotinic acid as a hepatoselective glucokinase activator clinical candidate for treating type 2 diabetes mellitus. J. Med. Chem. 55, 1318–1333 (2012)
Fujimoto, Y., Donahue, E. P. & Shiota, M. Defect in glucokinase translocation in Zucker diabetic fatty rats. Am. J. Physiol. Endocrinol. Metab. 287, E414–E423 (2004)
Shin, J. S., Torres, T. P., Catlin, R. L., Donahue, E. P. & Shiota, M. A defect in glucose-induced dissociation of glucokinase from the regulatory protein in Zucker diabetic fatty rats in the early stage of diabetes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R1381–R1390 (2007)
Choi, J. M., Seo, M. H., Kyeong, H. H., Kim, E. & Kim, H. S. Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase. Proc. Natl Acad. Sci. USA 110, 10171–10176 (2013)
Ashton, K. et al. Preparation of sulfonylpiperazine derivatives that interact with glucokinase regulatory protein for the treatment of diabetes and other diseases. US patent PCT Int. Appl. WO 2012027261. (2012)
Frenette, R. et al. Substituted 4-(2,2-diphenylethyl)pyridine-N-oxides as phosphodiesterase-4 inhibitors: SAR study directed toward the improvement of pharmacokinetic parameters. Bioorg. Med. Chem. Lett. 12, 3009–3013 (2002)
Hogan, P. J. & Cox, B. G. Aqueous process chemistry: the preparation of aryl sulfonyl chlorides. Org. Process Res. Dev. 13, 875–879 (2009)
Kreamer, B. L. et al. Use of a low-speed, iso-density percoll centrifugation method to increase the viability of isolated rat hepatocyte preparations. In Vitro Cell. Dev. Biol. 22, 201–211 (1986)
Acknowledgements
We would like to thank J. Calahan, J. Laubacher, M. Moore and D. Reid for pharmaceutics support, J. Civet for in vivo assistance, J. Han and R. Fachini for recombinant protein production, K. Kim for LC–MS/MS technical assistance, N. Nishimura and K. Yang for scale-up of AMG-3969, J. Chen for pharmacokinetic support, and A. Shaywitz and L. Rice for critical reading and editorial support of the manuscript.
Author information
Authors and Affiliations
Contributions
D.J.L., S.R.J., M.M.V. and C.H. designed experiments, analysed data and wrote the manuscript. K.M. performed SPR spectroscopy. D.J.S., K.S.A., K.L.A., L.D.P., C.F., L.L., M.D.B. and M.H.N. were responsible for the design and synthesis of AMG-1694 and AMG-3969. R.C. collected data and performed biochemical assays. M.C. developed and collected data for the hepatocyte GK translocation assay. J.W. validated and collected data for the hepatocyte 2DG uptake assay. K.C. and R.C.W. designed, and K.C. performed, the high-throughput screen. R.C.W. developed the GK–GKRP binding assay. M.W. designed experiments. M.V. and R.J.M.K. generated protein reagents. S.C., J.Z. and S.R.J. conducted crystallographic studies. G.V., E.J.G. and S.L.V. performed IHC. G.S., J.H. and R.K. conducted in vivo experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare competing financial interests as employees of Amgen Inc.
Extended data figures and tables
Extended Data Figure 1 Optimization of the initial high-throughput screen hit (left) to produce AMG-1694 (right).
IC50 values were determined using the AlphaScreen GK–GKRP binding assay.
Extended Data Figure 2 SPR and crystallographic studies of GK, GKRP, S6P and AMG1694.
a, The binding of GKRP to GK was determined by SPR spectroscopy, using GKRP-biotin immobilized on a streptavidin biosensor surface. Binding of untagged GK was monitored at a highest concentration of 0.125 μM. Kd = 0.005 ± 0.0003 μM; association rate constant (ka) = (2.7 × 105) ± (0.1 × 105) M−1 s−1; dissociation rate constant (kd) = (1.35 × 10−3) ± (0.01 × 10−3) s−1. b, Determination of the binding of S6P on GKRP. GKRP-biotin was immobilized at high density onto the SPR biosensor surface to allow small-molecule detection, and increasing concentrations of S6P (0.062, 0.185, 0.556, 1.667 and 5 μM indicated with arrows) were passed over the GKRP surface in the absence and presence of 50 µM S6P in the running buffer. S6P bound GKRP with a Kd = 0.029 ± 0.004 μM (ka = (8 × 104) ± (2 × 104) M−1s−1; kd = (2.2 × 10−3) ± (0.4 × 10−3) s−1), whereas no binding was observed when S6P was pre-bound, in agreement with the sugar binding pocket being blocked by S6P. c, d, Difference electron density maps for AMG-1694 (c) and S6P (d). Maps were calculated by 10 cycles of REFMAC (CCP4) refinement with both ligands removed from the calculations. Maps are contoured at ± 3σ. e, Ribbon drawing showing AMG-1694 in its binding site making hydrogen bonds to the backbone nitrogen of Ile 11 and the side chain of Arg 525. Illustrated using PyMOL (Schrödinger). f, Interaction map for AMG-1694 as generated by the program MOE (Chemical Computing Group). g, AMG-1694 requires residues 1–19 of GKRP for efficient binding. Immobilized biotinylated GKRP (20–625) on a biosensor surface was exposed to increasing concentrations of AMG-1694 (arrows; 0.247, 0.741, 2.2, 6.6 and 20 μM). Kd = 14 ± 0.4 μM; ka = (1.07 × 105) ± (0.02 × 105) M−1s−1; kd = 1.5 ± 0.1 s−1 (off-rate is outside of the range that can be measured accurately).
Extended Data Figure 3 GK translocation in a primary rat hepatocyte assay by glucose and AMG-1694.
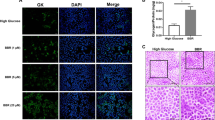
a, Hepatocytes incubated with increasing concentrations of glucose resulted in clear nuclear disappearance and cytoplasmic appearance of GK as detected by immunocytochemistry and visualized in pseudo colour using the ArrayScan platform. Scale bars, 50 μm. b, Image analysis of a permitted the GK nuclear/cytoplasmic difference to be calculated, illustrating a dose response with glucose exposure. c, GK translocation in a hepatocyte assay visualized in psuedo colour using an Operetta platform. Hepatocytes were incubated with increasing concentrations of AMG-1694, resulting in clear nuclear disappearance and cytoplasmic appearance of GK. Scale bars, 15 μm. d, Image analysis of c assessing the nuclear/cytoplasmic difference demonstrated GK translocation dose response with AMG-1694.
Extended Data Figure 4 Effects of AMG-1694 on GK translocation, insulin and triglyceride levels in Wistar and ZDF rats.
a, Wistar rats were dosed with AMG-1694, and 1 h later livers were analysed. Both 30 and 100 mg kg−1 doses elicited GK translocation that was greater than that observed with glucose/fructose (gluc/fruc; 2 g per 0.25 g per /kg). b, e, Insulin and triglyceride levels after 1 h in Wistar rats dosed with multiple doses of AMG-1694 as presented in a. c, f, Insulin and triglyceride levels in Wistar rats dosed with AMG-1694 as presented in Fig. 3b. d, g, Insulin and triglyceride levels in ZDF rats dosed with AMG-1694 as presented in Fig. 3c. Error bars denote s.e.m.; n = 3. *P < 0.05, ***P < 0.001 versus vehicle (ANOVA).
Extended Data Figure 5 A GK–GKRP disruptor piperazine derivative (AMG-3969).
a, Optimization of AMG-1694 (left) to produce AMG-3969 (right). b, Potency of AMG-3969 using GK translocation in a primary rat hepatocyte assay visualized using an Operetta platform.
Extended Data Figure 6 GK immunoblotting in AMG1694 dosed ZDF rats and GKRP–S6P cocrystal structure overlay with and without AMG-1694.
a, Expression of GK and GKRP levels were unaltered in livers of ZDF rats dosed with 100 mg kg−1 AMG-1694 for 4 days. The blots were stripped and reprobed with anti-β-actin antibody to ensure equal loading. b, The GKRP–S6P cocrystal structure overlay with and without AMG-1694. Crystals were originally grown as a complex of GKRP and S6P (shown in red). The crystals were then soaked in a solution containing AMG-1694 (shown in green).
Rights and permissions
About this article
Cite this article
Lloyd, D., St Jean, D., Kurzeja, R. et al. Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors. Nature 504, 437–440 (2013). https://doi.org/10.1038/nature12724
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12724
This article is cited by
-
GLUT9 as a potential drug target for chronic kidney disease: Drug target validation by a Mendelian randomization study
Journal of Human Genetics (2023)
-
Elevated circulating follistatin associates with an increased risk of type 2 diabetes
Nature Communications (2021)
-
A novel reverse two-hybrid method for the identification of missense mutations that disrupt protein–protein binding
Scientific Reports (2020)
-
Pharmacogenomic Studies of Current Antidiabetic Agents and Potential New Drug Targets for Precision Medicine of Diabetes
Diabetes Therapy (2020)
-
Non-alcoholic fatty liver disease and cardiovascular disease: assessing the evidence for causality
Diabetologia (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.