Abstract
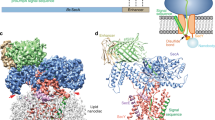
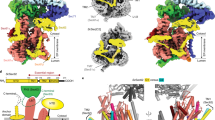
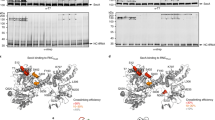
Many secretory proteins are targeted by signal sequences to a protein-conducting channel, formed by prokaryotic SecY or eukaryotic Sec61 complexes, and are translocated across the membrane during their synthesis1,2. Crystal structures of the inactive channel show that the SecY subunit of the heterotrimeric complex consists of two halves that form an hourglass-shaped pore with a constriction in the middle of the membrane and a lateral gate that faces the lipid phase3,4,5. The closed channel has an empty cytoplasmic funnel and an extracellular funnel that is filled with a small helical domain, called the plug. During initiation of translocation, a ribosome–nascent chain complex binds to the SecY (or Sec61) complex, resulting in insertion of the nascent chain. However, the mechanism of channel opening during translocation is unclear. Here we have addressed this question by determining structures of inactive and active ribosome–channel complexes with cryo-electron microscopy. Non-translating ribosome–SecY channel complexes derived from Methanocaldococcus jannaschii or Escherichia coli show the channel in its closed state, and indicate that ribosome binding per se causes only minor changes. The structure of an active E. coli ribosome–channel complex demonstrates that the nascent chain opens the channel, causing mostly rigid body movements of the amino- and carboxy-terminal halves of SecY. In this early translocation intermediate, the polypeptide inserts as a loop into the SecY channel with the hydrophobic signal sequence intercalated into the open lateral gate. The nascent chain also forms a loop on the cytoplasmic surface of SecY rather than entering the channel directly.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
Electron density maps have been submitted to the Electron Microscopy Data Bank (http://www.emdatabank.org/) under accession numbers EMD-5691, EMD-5692 and EMD-5693, and modelled structures to the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) under accession numbers 3J43, 3J44, 3J45, 3J46 and 1VVK.
References
Park, E. & Rapoport, T. A. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 (2012)
Shao, S. & Hegde, R. S. Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 27, 25–56 (2011)
Van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004)
Tsukazaki, T. et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455, 988–991 (2008)
Egea, P. F. & Stroud, R. M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl Acad. Sci. USA 107, 17182–17187 (2010)
Armache, J. P. et al. Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Res. 41, 1284–1293 (2013)
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008)
Ménétret, J. F. et al. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol. Cell 28, 1083–1092 (2007)
Ménétret, J. F. et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure 16, 1126–1137 (2008)
Becker, T. et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 (2009)
Frauenfeld, J. et al. Cryo-EM structure of the ribosome–SecYE complex in the membrane environment. Nature Struct. Mol. Biol. 18, 614–621 (2011)
Berk, V., Zhang, W., Pai, R. D. & Cate, J. H. D. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl Acad. Sci. USA 103, 15830–15834 (2006)
Zimmer, J., Nam, Y. & Rapoport, T. A. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 (2008)
Park, E. & Rapoport, T. A. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 473, 239–242 (2011)
Park, E. & Rapoport, T. A. Bacterial protein translocation requires only one copy of the SecY complex in vivo . J. Cell Biol. 198, 881–893 (2012)
Schierle, C. F. et al. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185, 5706–5713 (2003)
Nakatogawa, H. & Ito, K. The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636 (2002)
Zhang, Y. et al. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli . Mol. Cell 12, 913–923 (2003)
Muto, H., Nakatogawa, H. & Ito, K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell 22, 545–552 (2006)
Sengupta, J., Agrawal, R. K. & Frank, J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl Acad. Sci. USA 98, 11991–11996 (2001)
Lycklama a Nijeholt, J. A., Bulacu, M., Marrink, S. J. & Driessen, A. J. Immobilization of the plug domain inside the SecY channel allows unrestricted protein translocation. J. Biol. Chem. 285, 23747–23754 (2010)
Plath, K., Mothes, W., Wilkinson, B. M., Stirling, C. J. & Rapoport, T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94, 795–807 (1998)
Knyazev, D. G. et al. The bacterial translocon SecYEG opens upon ribosome binding. J. Biol. Chem. 288, 17941–17946 (2013)
Shaw, A. S., Rottier, P. J. & Rose, J. K. Evidence for the loop model of signal-sequence insertion into the endoplasmic reticulum. Proc. Natl Acad. Sci. USA 85, 7592–7596 (1988)
Hizlan, D. et al. Structure of the SecY complex unlocked by a preprotein mimic. Cell Rep 1, 21–28 (2012)
Matlack, K. E., Misselwitz, B., Plath, K. & Rapoport, T. A. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97, 553–564 (1999)
Nicchitta, C. V. & Blobel, G. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell 73, 989–998 (1993)
Tsukazaki, T. et al. Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238 (2011)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000)
Alami, M., Dalal, K., Lelj-Garolla, B., Sligar, S. G. & Duong, F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004 (2007)
Nath, A., Atkins, W. M. & Sligar, S. G. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 (2007)
Cannon, K. S., Or, E., Clemons, W. M., Jr, Shibata, Y. & Rapoport, T. A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 169, 219–225 (2005)
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999)
Jenner, L. B., Demeshkina, N., Yusupova, G. & Yusupov, M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nature Struct. Mol. Biol. 17, 555–560 (2010)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
DiMaio, F., Tyka, M. D., Baker, M. L., Chiu, W. & Baker, D. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 392, 181–190 (2009)
Acknowledgements
We thank K. Matlack and T. Guettler for reading the manuscript. This work was supported by National Institutes of Health grants GM067887 to J.C.G., GM080139 to S.J.L., GM052586 to T.A.R. and GM45377 to C.W.A. T.A.R. is a Howard Hughes Institute investigator.
Author information
Authors and Affiliations
Contributions
E.P. designed and purified RNC–channel complexes, J.-F.M. and C.W.A. obtained and analysed the electron microscopy data, J.C.G. helped with MDFF and channel modelling, S.J.L. helped with data analysis, W.L. and A.W. purified M. jannaschii components, and T.A.R., E.P. and C.W.A. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-22 and Supplementary Tables 1-3. (PDF 15975 kb)
Conformational changes in the E. coli SecY channel during translocation - part I
The video illustrates conformational changes in the SecY channel that occur when a nascent chain with a signal sequence is bound in a looped configuration. The nascent chain has been omitted for clarity. A morph was generated between models of the closed and active SecY complex in Chimera, with the channel viewed from the front. The N-terminal half of SecY is in light blue, the C-terminal half is in red, SecE is dark blue, SecG is brown, and the plug is yellow. Six isoleucines that line the pore are shown as ball and stick representations in dark grey. Ribosomal helices H6, H7, H50 and H50 are shown in white; proteins L23, and L24 are green, and L29 is dark grey. The position and conformation of the 6/7 loop was fixed during the simulation. Note that SecE tilts to accommodate movements of the two halves of SecY, and helix 8b moves downwards towards the lateral gate, while helix 9 remains anchored to the large ribosomal subunit. These movements open the lateral gate between the two halves of SecY. (MOV 5307 kb)
Conformational changes in the E. coli SecY channel during translocation - part II
The video illustrates conformational changes in SecY channel that occur when a nascent chain with a signal sequence is bound in a looped configuration. The channel is viewed from the top. The nascent chain has been omitted for clarity. Note that the N-terminal half of SecY rotates and tilts backwards, approximating a rigid body movement that also involves SecG. However, helix 3 bends and the junction between helix 5 and 6 also moves to accommodate these large movements. These movements create a large translocation pore with access to the opened lateral gate. (MOV 5618 kb)
Visualizing the nascent chain in the active E. coli SecY channel
The video illustrates the orientation of a closed SecY channel beneath the ribosome and then shows a progression from a closed to an open SecY. The nascent chain containing the signal sequence helix (residues 1-73; in green) is bound in a looped configuration and a cytoplasmic loop resides in a V-shaped canyon bounded by the 6/7 connection and helix 10. The N-terminal half of SecY is in light blue, the C-terminal half is in red, SecE is dark blue, SecG is brown, and the plug is yellow. Six isoleucines that line the pore are shown as ball and stick representations in dark grey. Ribosomal helices H6, H7, H50 and H59 are shown in white; ribosomal proteins are colored as follows: L23 (purple), L29 (dark grey) and L29 (yellow). The extended nascent chain exiting the tunnel couldinteract with the 6/7 loop to help stabilize the translocating ribosomechannel complex. (MOV 12322 kb)
Rights and permissions
About this article
Cite this article
Park, E., Ménétret, JF., Gumbart, J. et al. Structure of the SecY channel during initiation of protein translocation. Nature 506, 102–106 (2014). https://doi.org/10.1038/nature12720
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12720
This article is cited by
-
Dynamic coupling of fast channel gating with slow ATP-turnover underpins protein transport through the Sec translocon
The EMBO Journal (2023)
-
A common mechanism of Sec61 translocon inhibition by small molecules
Nature Chemical Biology (2023)
-
GREACE-assisted adaptive laboratory evolution in endpoint fermentation broth enhances lysine production by Escherichia coli
Microbial Cell Factories (2019)
-
The Principles of Protein Targeting and Transport Across Cell Membranes
The Protein Journal (2019)
-
Structural Basis of the Sec Translocon and YidC Revealed Through X-ray Crystallography
The Protein Journal (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.