Abstract
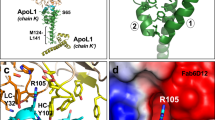
Members of the CD36 superfamily of scavenger receptor proteins are important regulators of lipid metabolism and innate immunity. They recognize normal and modified lipoproteins, as well as pathogen-associated molecular patterns. The family consists of three members: SR-BI (which delivers cholesterol to the liver and steroidogenic organs and is a co-receptor for hepatitis C virus), LIMP-2/LGP85 (which mediates lysosomal delivery of β-glucocerebrosidase and serves as a receptor for enterovirus 71 and coxsackieviruses) and CD36 (a fatty-acid transporter and receptor for phagocytosis of effete cells and Plasmodium-infected erythrocytes). Notably, CD36 is also a receptor for modified lipoproteins and β-amyloid, and has been implicated in the pathogenesis of atherosclerosis and of Alzheimer’s disease1. Despite their prominent roles in health and disease, understanding the function and abnormalities of the CD36 family members has been hampered by the paucity of information about their structure. Here we determine the crystal structure of LIMP-2 and infer, by homology modelling, the structure of SR-BI and CD36. LIMP-2 shows a helical bundle where β-glucocerebrosidase binds, and where ligands are most likely to bind to SR-BI and CD36. Remarkably, the crystal structure also shows the existence of a large cavity that traverses the entire length of the molecule. Mutagenesis of SR-BI indicates that the cavity serves as a tunnel through which cholesterol(esters) are delivered from the bound lipoprotein to the outer leaflet of the plasma membrane. We provide evidence supporting a model2 whereby lipidic constituents of the ligands attached to the receptor surface are handed off to the membrane through the tunnel, accounting for the selective lipid transfer characteristic of SR-BI and CD36.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Canton, J., Neculai, D. & Grinstein, S. Scavenger receptors in homeostasis and immunity. Nature Rev. Immunol. 13, 621–634 (2013)
Rodrigueza, W. V. et al. Mechanism of scavenger receptor class B type I-mediated selective uptake of cholesteryl esters from high density lipoprotein to adrenal cells. J. Biol. Chem. 274, 20344–20350 (1999)
Yu, M. et al. Exoplasmic cysteine Cys384 of the HDL receptor SR-BI is critical for its sensitivity to a small-molecule inhibitor and normal lipid transport activity. Proc. Natl Acad. Sci. USA 108, 12243–12248 (2011)
Rasmussen, J. T., Berglund, L., Rasmussen, M. S. & Petersen, T. E. Assignment of disulfide bridges in bovine CD36. Eur. J. Biochem. 257, 488–494 (1998)
Reczek, D. et al. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of β-glucocerebrosidase. Cell 131, 770–783 (2007)
Blanz, J. et al. Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand β-glucocerebrosidase. Hum. Mol. Genet. 19, 563–572 (2010)
Zachos, C., Blanz, J., Saftig, P. & Schwake, M. A critical histidine residue within LIMP-2 mediates pH sensitive binding to its ligand β-glucocerebrosidase. Traffic 13, 1113–1123 (2012)
Ryeom, S. W., Silverstein, R. L., Scotto, A. & Sparrow, J. R. Binding of anionic phospholipids to retinal pigment epithelium may be mediated by the scavenger receptor CD36. J. Biol. Chem. 271, 20536–20539 (1996)
Jimenez-Dalmaroni, M. J. et al. Soluble CD36 ectodomain binds negatively charged diacylglycerol ligands and acts as a co-receptor for TLR2. PLoS ONE 4, e7411 (2009)
Puente Navazo, M. D., Daviet, L., Ninio, E. & McGregor, J. L. Identification on human CD36 of a domain (155-183) implicated in binding oxidized low-density lipoproteins (Ox-LDL). Arterioscler. Thromb. Vasc. Biol. 16, 1033–1039 (1996)
Stylianou, I. M. et al. Novel ENU-induced point mutation in scavenger receptor class B, member 1, results in liver specific loss of SCARB1 protein. PLoS ONE 4, e6521 (2009)
Chadwick, A. C. & Sahoo, D. Functional characterization of newly-discovered mutations in human SR-BI. PLoS ONE 7, e45660 (2012)
Kar, N. S., Ashraf, M. Z., Valiyaveettil, M. & Podrez, E. A. Mapping and characterization of the binding site for specific oxidized phospholipids and oxidized low density lipoprotein of scavenger receptor CD36. J. Biol. Chem. 283, 8765–8771 (2008)
Rigotti, A., Miettinen, H. E. & Krieger, M. The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocr. Rev. 24, 357–387 (2003)
Gu, X. et al. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J. Biol. Chem. 273, 26338–26348 (1998)
Liu, B. & Krieger, M. Highly purified scavenger receptor class B, type I reconstituted into phosphatidylcholine/cholesterol liposomes mediates high affinity high density lipoprotein binding and selective lipid uptake. J. Biol. Chem. 277, 34125–34135 (2002)
Gu, X., Kozarsky, K. & Krieger, M. Scavenger receptor class B, type I-mediated [3H]cholesterol efflux to high and low density lipoproteins is dependent on lipoprotein binding to the receptor. J. Biol. Chem. 275, 29993–30001 (2000)
Ji, Y. et al. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J. Biol. Chem. 272, 20982–20985 (1997)
Nieland, T. J., Penman, M., Dori, L., Krieger, M. & Kirchhausen, T. Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI. Proc. Natl Acad. Sci. USA 99, 15422–15427 (2002)
Nieland, T. J. et al. Identification of the molecular target of small molecule inhibitors of HDL receptor SR-BI activity. Biochemistry 47, 460–472 (2008)
Yu, M., Lau, T. Y., Carr, S. A. & Krieger, M. Contributions of a disulfide bond and a reduced cysteine side chain to the intrinsic activity of the high-density lipoprotein receptor SR-BI. Biochemistry 51, 10044–10055 (2012)
Acknowledgements
We thank W. Temple, D. Cossar, S. Graslund, C. Arrowsmith, R. Stahelin, L. Andresen, J. Groth, M. Langer and J. Scott for assistance and discussions. This study was supported by the Canadian Institutes for Health Research (grants MOP-102474 and MOP-126069) and the Deutsche Forschungsgemeinschaft (grants GRK1459 to M.S. and SFB877). F.Z. is supported through the Böhringer Ingelheim Fonds. W.S.T. is the recipient of a Canada Research Chair in Molecular Cell Biology. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canada Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.G., W.S.T., P.S., M.S., S.D.-P. and D.N. designed the research; D.N., M.R., M.N., P.L., A.S., J.C.P., R.C., J. Plumb, F.Z. and J. Peters performed the experiments; S.G., W.S.T., P.S., M.S., S.D.-P., D.N. and F.Z. analysed the results; S.G. wrote the paper; W.S.T., P.S., M.S., S.D.-P., D.N., J. Peters and F.Z. edited and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary Figures 1-12, Supplementary Table 1 and Additional references. (PDF 22872 kb)
Rights and permissions
About this article
Cite this article
Neculai, D., Schwake, M., Ravichandran, M. et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature 504, 172–176 (2013). https://doi.org/10.1038/nature12684
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12684
This article is cited by
-
The computational analyses, molecular dynamics of fatty-acid transport mechanism to the CD36 receptor
Scientific Reports (2021)
-
Novel Functions of Endothelial Scavenger Receptor Class B Type I
Current Atherosclerosis Reports (2021)
-
Cellular receptors for enterovirus A71
Journal of Biomedical Science (2020)
-
Molecular determinants of SR-B1-dependent Plasmodium sporozoite entry into hepatocytes
Scientific Reports (2020)
-
Lipid Uptake by Alveolar Macrophages Drives Fibrotic Responses to Silica Dust
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.