Abstract
Brown adipose tissue (BAT) dissipates chemical energy in the form of heat as a defence against hypothermia and obesity. Current evidence indicates that brown adipocytes arise from Myf5+ dermotomal precursors through the action of PR domain containing protein 16 (PRDM16) transcriptional complex1,2. However, the enzymatic component of the molecular switch that determines lineage specification of brown adipocytes remains unknown. Here we show that euchromatic histone-lysine N-methyltransferase 1 (EHMT1) is an essential BAT-enriched lysine methyltransferase in the PRDM16 transcriptional complex and controls brown adipose cell fate. Loss of EHMT1 in brown adipocytes causes a severe loss of brown fat characteristics and induces muscle differentiation in vivo through demethylation of histone 3 lysine 9 (H3K9me2 and 3) of the muscle-selective gene promoters. Conversely, EHMT1 expression positively regulates the BAT-selective thermogenic program by stabilizing the PRDM16 protein. Notably, adipose-specific deletion of EHMT1 leads to a marked reduction of BAT-mediated adaptive thermogenesis, obesity and systemic insulin resistance. These data indicate that EHMT1 is an essential enzymatic switch that controls brown adipose cell fate and energy homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Seale, P. et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967 (2008)
Kajimura, S. et al. Initiation of myoblast to brown fat switch by a PRDM16–C/EBP-β transcriptional complex. Nature 460, 1154–1158 (2009)
Nedergaard, J., Bengtsson, T. & Cannon, B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. 293, E444–E452 (2007)
van Marken Lichtenbelt, W. D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009)
Saito, M. et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531 (2009)
Virtanen, K. A. et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360, 1518–1525 (2009)
Atit, R. et al. β-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev. Biol. 296, 164–176 (2006)
Timmons, J. A. et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl Acad. Sci. USA 104, 4401–4406 (2007)
Kajimura, S., Seale, P. & Spiegelman, B. M. Transcriptional control of brown fat development. Cell Metab. 11, 257–262 (2010)
Shing, D. C. et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J. Clin. Invest. 117, 3696–3707 (2007)
Pinheiro, I. et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948–960 (2012)
Tachibana, M. et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev. 19, 815–826 (2005)
Kleefstra, T. et al. Disruption of the gene euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J. Med. Genet. 42, 299–306 (2005)
Cormier-Daire, V. et al. Cryptic terminal deletion of chromosome 9q34: a novel cause of syndromic obesity in childhood? J. Med. Genet. 40, 300–303 (2003)
Willemsen, M. H. et al. Update on Kleefstra syndrome. Mol. Syndromol. 2, 202–212 (2012)
Kajimura, S. et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 22, 1397–1409 (2008)
Schaefer, A. et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64, 678–691 (2009)
Tachibana, M., Matsumura, Y., Fukuda, M., Kimura, H. & Shinkai, Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 27, 2681–2690 (2008)
Eguchi, J. et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13, 249–259 (2011)
Almind, K., Manieri, M., Sivitz, W. I., Cinti, S. & Kahn, C. R. Ectopic brown adipose tissue in muscle provides a mechanism for differences in risk of metabolic syndrome in mice. Proc. Natl Acad. Sci. USA 104, 2366–2371 (2007)
Cannon, B. & Nedergaard, J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. J. Exp. Biol. 214, 242–253 (2011)
Ouellet, V. et al. Outdoor temperature, age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. J. Clin. Endocrinol. Metab. 96, 192–199 (2012)
Wu, Q. et al. Fatty acid transport protein 1 is required for nonshivering thermogenesis in brown adipose tissue. Diabetes 55, 3229–3237 (2006)
Feldmann, H. M., Golozoubova, V., Cannon, B. & Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209 (2009)
Yoneshiro, T. et al. Impact of UCP1 and beta3AR gene polymorphisms on age-related changes in brown adipose tissue and adiposity in humans. Int. J. Obes. 37, 993–998 (2013)
Huh, M. S., Parker, M. H., Scime, A., Parks, R. & Rudnicki, M. A. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 166, 865–876 (2004)
Mao, X. et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nature Cell Biol. 8, 516–523 (2006)
Liisberg Aune, U., Ruiz, L. & Kajimura, S. Isolation and differentiation of stromal vascular cells to beige/brite cells. J Visual. Exp. e50191 (2013)
Takahashi, A. et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol. Cell 45, 123–131 (2012)
Kurn, N. et al. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 51, 1973–1981 (2005)
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnol. 31, 46–53 (2013)
Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Kersey, P. J. et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004)
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods 4, 207–214 (2007)
Seale, P. et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 6, 38–54 (2007)
Ohno, H., Shinoda, K., Spiegelman, B. M. & Kajimura, S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 15, 395–404 (2012)
Acknowledgements
We are grateful to A. Tarakhovsky, E. D. Rosen, Y. Shinkai and E. Hara for providing mice and plasmids. We thank our colleagues in the University of California, San Francisco, including Y. Qiu, A. Chawla, C. Paillart, S. Koliwad, M. Robblee, D. Scheel, S. Ohata, L. Mera, D. Lowe, S. Sonne, S. Keylin, I. Luijten, H. Hong and E. Tomoda for their assistance. This work was supported by grants from the National Institutes of Health (DK087853 and DK97441) to S.K. We acknowledge supports from the DERC center grant (DK63720), University of California, San Francisco Program for Breakthrough Biomedical Research program, the Pew Charitable Trust, and PRESTO from the Japan Science and Technology Agency to S.K. H.O. is supported by the Manpei Suzuki Diabetes Foundation. K.S. is supported by a fellowship from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Contributions
S.K. and H.O. conceived and designed the experiments. All authors performed the experiments and analysed the data. S.K. and H.O. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
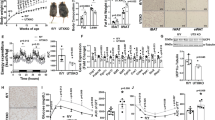
Extended Data Figure 1 EHMT1 regulates endogenous PRDM16 protein expression in vivo.
a, The putative BAT was micro-dissected from WT and Ehmt1myf5 knockout embryos. mRNA expression of Prdm16 was measured by qRT–PCR. Data are presented as mean and s.e.m. (n = 8–10). b, Western blotting to detect endogenous EHMT1, PRDM16, UCP1 and MHC in BAT from WT and Ehmt1myf5 knockout embryos. α-Tublin protein was shown as a loading control.
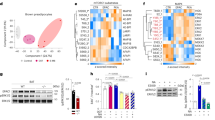
Extended Data Figure 2 Ectopic activation of skeletal-muscle-selective genes and reduction of BAT-selective genes in the BAT from Ehmt1adipo knockout mice.
a, Western blotting for endogenous EHMT1 in BAT and liver from WT and Ehmt1adipo knockout mice. β-Actin protein was shown as a loading control. b, Amounts of mRNA expression of BAT, skeletal muscle, white fat and beige-fat selective genes in BAT from Ehmt1adipo knockout mice. Values were normalized to those in WT mice. The amounts of mRNA were visualized by a heat-map using Multi Experiment Viewer. c, Venn diagram showing the overlapped genes between Ehmt1myf5 knockout and Ehmt1adipo knockout mice. RNA-sequencing and gene ontology analyses identified 33 genes that were similarly dysregulated both in the Ehmt1myf5 knockout BAT and the Ehmt1adipo knockout BAT. The mRNA expression values were normalized to WT mice for each knockout model and visualized by a heat-map using Multi Experiment Viewer. The colour scale shows the amounts of mRNA of the genes in a blue (low)–white (no change)–red (high) scheme.
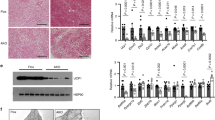
Extended Data Figure 3 EHMT1 is required for beige/brite cell development.
a, The b3-AR agonist CL316,243 at a dose of 0.5 mg kg−1 or saline were administered to WT or Ehmt1adipo knockout mice for 7 days. Inguinal WAT was collected for gene expression analysis. Amounts of mRNA expression of BAT and beige-fat selective genes (as indicated) were measured by qRT–PCR (n = 3–6). †Significant between saline and CL316,243 in WT mice. b, Immunohistochemistry for UCP1 in a. Scale bar, 100 μm. Nuclei were stained with DAPI. c, To test a cell-autonomous requirement for EHMT1 in beige/brite cell development, the stromal vascular (SV) fractions were isolated from the inguinal WAT of Ehmt1flox/flox mice. Cells were infected with adenovirus expressing GFP or Cre. The SV cells were differentiated in the presence or absence of rosiglitazone (Rosi) at 0.5 μM. Amounts of mRNA expression of BAT-selective genes (as indicated) were measured by qRT–PCR. Deletion of Ehmt1 was confirmed by qRT–PCR (right graph) (n = 3); data are presented as mean and s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-23 and Supplementary Tables 1-2. (PDF 1902 kb)
Rights and permissions
About this article
Cite this article
Ohno, H., Shinoda, K., Ohyama, K. et al. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504, 163–167 (2013). https://doi.org/10.1038/nature12652
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12652
This article is cited by
-
Association between single nucleotide polymorphisms, TGF-β1 promoter methylation, and polycystic ovary syndrome
BMC Pregnancy and Childbirth (2024)
-
The rs1421085 variant within FTO promotes brown fat thermogenesis
Nature Metabolism (2023)
-
Linker histone variant H1.2 is a brake on white adipose tissue browning
Nature Communications (2023)
-
Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies
Signal Transduction and Targeted Therapy (2023)
-
Overlapping pathogenic de novo CNVs in neurodevelopmental disorders and congenital anomalies impacting constraint genes regulating early development
Human Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.