Abstract
Animal cells harbour multiple innate effector mechanisms that inhibit virus replication. For the pathogenic retrovirus human immunodeficiency virus type 1 (HIV-1), these include widely expressed restriction factors1, such as APOBEC3 proteins2, TRIM5-α3, BST2 (refs 4, 5) and SAMHD1 (refs 6, 7), as well as additional factors that are stimulated by type 1 interferon (IFN)8,9,10,11,12,13,14. Here we use both ectopic expression and gene-silencing experiments to define the human dynamin-like, IFN-induced myxovirus resistance 2 (MX2, also known as MXB) protein as a potent inhibitor of HIV-1 infection and as a key effector of IFN-α-mediated resistance to HIV-1 infection. MX2 suppresses infection by all HIV-1 strains tested, has equivalent or reduced effects on divergent simian immunodeficiency viruses, and does not inhibit other retroviruses such as murine leukaemia virus. The Capsid region of the viral Gag protein dictates susceptibility to MX2, and the block to infection occurs at a late post-entry step, with both the nuclear accumulation and chromosomal integration of nascent viral complementary DNA suppressed. Finally, human MX1 (also known as MXA), a closely related protein that has long been recognized as a broadly acting inhibitor of RNA and DNA viruses, including the orthomyxovirus influenza A virus15,16, does not affect HIV-1, whereas MX2 is ineffective against influenza virus. MX2 is therefore a cell-autonomous, anti-HIV-1 resistance factor whose purposeful mobilization may represent a new therapeutic approach for the treatment of HIV/AIDS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Malim, M. H. & Bieniasz, P. D. HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2, a006940 (2012)
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002)
Stremlau, M. et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004)
Neil, S. J., Zang, T. & Bieniasz, P. D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008)
Van Damme, N. et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 (2008)
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011)
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011)
Asmuth, D. M. et al. Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa-2a in HIV-1-monoinfected participants: a phase II clinical trial. J. Infect. Dis. 201, 1686–1696 (2010)
Bitzegeio, J., Sampias, M., Bieniasz, P. D. & Hatziioannou, T. Adaptation to the interferon-induced antiviral state by human and simian immunodeficiency viruses. J. Virol. 87, 3549–3560 (2013)
Goujon, C. & Malim, M. H. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J. Virol. 84, 9254–9266 (2010)
Ho, D. D. et al. Recombinant human interferon alfa-A suppresses HTLV-III replication in vitro. Lancet 325, 602–604 (1985)
Meylan, P. R., Guatelli, J. C., Munis, J. R., Richman, D. D. & Kornbluth, R. S. Mechanisms for the inhibition of HIV replication by interferons-α, -β, and -γ in primary human macrophages. Virology 193, 138–148 (1993)
Pillai, S. K. et al. Role of retroviral restriction factors in the interferon-α-mediated suppression of HIV-1 in vivo. Proc. Natl Acad. Sci. USA 109, 3035–3040 (2012)
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011)
Haller, O. & Kochs, G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. 31, 79–87 (2011)
Pavlovic, J., Zurcher, T., Haller, O. & Staeheli, P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 64, 3370–3375 (1990)
Goujon, C. et al. Evidence for IFNα-induced, SAMHD1-independent inhibitors of early HIV-1 infection. Retrovirology 10, 23 (2013)
Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573 (2004)
Kim, B. H., Shenoy, A. R., Kumar, P., Bradfield, C. J. & MacMicking, J. D. IFN-inducible GTPases in host cell defense. Cell Host Microbe 12, 432–444 (2012)
Gao, S. et al. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35, 514–525 (2011)
Mitchell, P. S. et al. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe 12, 598–604 (2012)
King, M. C., Raposo, G. & Lemmon, M. A. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc. Natl Acad. Sci. USA 101, 8957–8962 (2004)
Melén, K. et al. Human MxB protein, an interferon-α-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 271, 23478–23486 (1996)
Brass, A. L. et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 (2008)
König, R. et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135, 49–60 (2008)
Lee, K. et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010)
Shah, V. B. et al. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 87, 422–432 (2013)
Pitossi, F. et al. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J. Virol. 67, 6726–6732 (1993)
Ponten, A., Sick, C., Weeber, M., Haller, O. & Kochs, G. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J. Virol. 71, 2591–2599 (1997)
Brass, A. L. et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 (2009)
Adachi, A. et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59, 284–291 (1986)
Fouchier, R. A., Meyer, B. E., Simon, J. H., Fischer, U. & Malim, M. H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16, 4531–4539 (1997)
Simon, J. H., Southerling, T. E., Peterson, J. C., Meyer, B. E. & Malim, M. H. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 69, 4166–4172 (1995)
Schindler, M. et al. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77, 10548–10556 (2003)
Ochsenbauer, C. et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 86, 2715–2728 (2012)
Ryan-Graham, M. A. & Peden, K. W. Both virus and host components are important for the manifestation of a Nef− phenotype in HIV-1 and HIV-2. Virology 213, 158–168 (1995)
Sauter, D. et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421 (2009)
Soares, M. A. et al. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228, 394–399 (1997)
Sakai, H. et al. Genetic characterization of simian immunodeficiency virus isolated from an African mandrill. Arch. Virol. 125, 1–14 (1992)
Gaddis, N. C. et al. Further investigation of simian immunodeficiency virus Vif function in human cells. J. Virol. 78, 12041–12046 (2004)
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996)
Mangeot, P. E. et al. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol. Ther. 5, 283–290 (2002)
Saenz, D. T., Teo, W., Olsen, J. C. & Poeschla, E. M. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5α proteins. J. Virol. 79, 15175–15188 (2005)
O'Rourke, J. P., Newbound, G. C., Kohn, D. B., Olsen, J. C. & Bunnell, B. A. Comparison of gene transfer efficiencies and gene expression levels achieved with equine infectious anemia virus- and human immunodeficiency virus type 1-derived lentivirus vectors. J. Virol. 76, 1510–1515 (2002)
Jarrosson-Wuilleme, L. et al. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 80, 1152–1159 (2006)
Schaller, T. et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7, e1002439 (2011)
Cordeil, S. et al. Evidence for a different susceptibility of primate lentiviruses to type I interferons. J. Virol. 87, 2587–2596 (2013)
Pear, W. S. et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92, 3780–3792 (1998)
Mangeot, P. E. et al. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Ther. 19, 1656–1666 (2011)
Wei, X. et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905 (2002)
Shi, W., Oshlack, A. & Smyth, G. K. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 38, e204 (2010)
Markusic, D., Oude-Elferink, R., Das, A. T., Berkhout, B. & Seppen, J. Comparison of single regulated lentiviral vectors with rtTA expression driven by an autoregulatory loop or a constitutive promoter. Nucleic Acids Res. 33, e63 (2005)
Trichas, G., Begbie, J. & Srinivas, S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 6, 40 (2008)
Donnelly, M. L. et al. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J. Gen. Virol. 82, 1013–1025 (2001)
Bamming, D. & Horvath, C. M. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284, 9700–9712 (2009)
Yee, J. K. et al. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl Acad. Sci. USA 91, 9564–9568 (1994)
Cauldwell, A. V., Moncorge, O. & Barclay, W. S. Unstable polymerase-nucleoprotein interaction is not responsible for avian influenza virus polymerase restriction in human cells. J. Virol. 87, 1278–1284 (2013)
Neumann, G. & Hobom, G. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76, 1709–1717 (1995)
Pertel, T. et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011)
Newman, E. N. et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15, 166–170 (2005)
O’Doherty, U., Swiggard, W. J., Jeyakumar, D., McGain, D. & Malim, M. H. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76, 10942–10950 (2002)
Acknowledgements
We wish to thank L. Apolonia, A. Cimarelli, B. Hahn, T. Hatziioannou, J. Kappes, P. Mangeot, S. Papaioannou, N. Parrish, N. Sherer and C. Swanson for the provision of reagents and for discussions, and M. Mirza, E. Papouli, Q. Oscar Y. Li and G. Pacini for technical assistance. This work was supported by the UK Medical Research Council, the National Institutes of Health (DA033773), the European Commission’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. PIEF-GA-2009-237501 (to C.G.), a Wellcome Trust Research Training Fellowship (to T.D.) and the Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust.
Author information
Authors and Affiliations
Contributions
C.G. and M.H.M. designed the study and wrote the manuscript. C.G. carried out the experiments. O.M. and W.S.B. designed and carried out the influenza A virus experiment. O.M., H.B., T.D., C.C.W., T.S. and S.H. provided technical assistance. R.S. performed the microarray analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
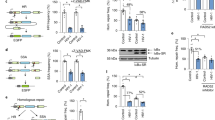
Extended Data Figure 1 MX2 inhibits infection with VSV-G-pseudotyped HIV-1.
U87-MG CD4+ CXCR4+ cells were transduced with EasiLV expressing the different candidate cDNAs, CD8, GFP or TRIMCyp cDNA controls and treated with doxycycline for 48 h, left untransduced (Ctrl) or treated with IFN-α for 24 h before infection. The cells were infected with increasing viral inputs of VSV-G-pseudotyped NL4-3/Nef-IRES-Renilla (0.04–5 ng p24Gag) and infection efficiency was monitored at 48 h by measuring Renilla activity. Mean relative infection efficiencies from four independent experiments are shown.
Extended Data Figure 2 MX2 is active in other cell types.
a, CEM-SS cells were transduced with EasiLV expressing CD8 or MX2 and treated with doxycycline for 4–5 days and infected with increasing viral inputs of VSV-G-pseudotyped NL4-3/Nef-IRES-GFP (0.2–25 ng p24Gag). The efficiency of infection was measured at 72 h by scoring the percentage of GFP-expressing cells, among the E2-Crimson-expressing cells, by flow cytometry. Mean percentages of infected cells from independent duplicates are shown. b, 293T cells were transduced with lentiviral vectors containing a bicistronic expression cassette for either CD8 or MX2 with the puromycinR gene, and selected for puromycin resistance. Transduced cells were infected with increasing viral inputs of VSV-G-pseudotyped NL4-3/Nef-IRES-Renilla (0.2–25 ng p24Gag) and Renilla activity was measured at 48 h. Mean relative infection efficiencies from three independent experiments are shown.
Extended Data Figure 3 MX2 relative levels of expression and IFN-α-inducibility.
RNA was extracted from control (Ctrl) or IFN-α-treated (IFNα) PMA-treated or dividing THP-1 cells, MDMs, primary CD4+ T cells, U87-MG, HT1080, CEM, CEM-SS, HUT78 and Jurkat cells. 500-ng aliquots of RNA were reverse transcribed and the levels of MX2, as well as ISG15, and two endogenous controls, GAPDH and β-actin (ACTB), were analysed by qRT–PCR. The graph shows the mean of relative levels of expression (normalized to both endogenous controls and compared to THP-1 without IFN-α treatment) obtained in three independent experiments.
Extended Data Figure 4 MX2 participates in IFN-α-induced resistance to HIV-1 in monocytic THP-1 cells.
a, THP-1 cells expressing a control shRNA or two different shRNAs targeting MX2 were treated with or without IFN-α (500 U ml−1) for 24 h. Cells were infected with five different doses of NL4-3/Nef-IRES-Renilla (0.04–25 ng p24Gag) for 48 h, and Renilla activity was measured. Mean relative infection efficiencies from two independent experiments are shown. b, Immunoblot analysis of parallel samples from a. Protein levels of MX2 and APOBEC3G (A3G, positive control for IFN-α treatment) were determined, and tubulin served as a loading control.
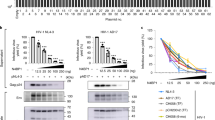
Extended Data Figure 5 MX2 blocks productive HIV-1IIIB infection.
U87-MG CD4+ CXCR4+ cells were transduced with EasiLV expressing CD8 or MX2 and treated with doxycycline for 48 h, left untransduced (Ctrl) or treated with IFN-α for 24 h before infection. Increasing viral inocula (indicated in ng p24Gag) were used to infect the cells with HIV-1IIIB. The levels of infection were analysed at 48 h by measuring the percentage of cells expressing p24Gag after intracellular staining. The means of three independent experiments are shown. This analysis accompanies the experiments shown in Fig. 3.
Extended Data Figure 6 Expression of wild-type and GTPase domain mutants of MX1 and MX2.
Immunoblot analysis of parallel samples from Fig. 4e. Protein levels of Flag-tagged MX1 and MX2 proteins were determined using a Flag-specific antibody and tubulin served as a loading control.
Rights and permissions
About this article
Cite this article
Goujon, C., Moncorgé, O., Bauby, H. et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502, 559–562 (2013). https://doi.org/10.1038/nature12542
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12542
This article is cited by
-
Emerging roles of the Protein Phosphatase 1 (PP1) in the context of viral infections
Cell Communication and Signaling (2024)
-
Innate immune regulation in HIV latency models
Retrovirology (2022)
-
The Hippo signaling component LATS2 enhances innate immunity to inhibit HIV-1 infection through PQBP1-cGAS pathway
Cell Death & Differentiation (2022)
-
HIV-1 capsid variability: viral exploitation and evasion of capsid-binding molecules
Retrovirology (2021)
-
Arginyl-tRNA-protein transferase 1 contributes to governing optimal stability of the human immunodeficiency virus type 1 core
Retrovirology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.