Abstract
Down’s syndrome results from full or partial trisomy of chromosome 21. However, the consequences of the underlying gene–dosage imbalance on adult tissues remain poorly understood. Here we show that in Ts65Dn mice, which are trisomic for 132 genes homologous to genes on human chromosome 21, triplication of Usp16 reduces the self-renewal of haematopoietic stem cells and the expansion of mammary epithelial cells, neural progenitors and fibroblasts. In addition, Usp16 is associated with decreased ubiquitination of Cdkn2a and accelerated senescence in Ts65Dn fibroblasts. Usp16 can remove ubiquitin from histone H2A on lysine 119, a critical mark for the maintenance of multiple somatic tissues. Downregulation of Usp16, either by mutation of a single normal Usp16 allele or by short interfering RNAs, largely rescues all of these defects. Furthermore, in human tissues overexpression of USP16 reduces the expansion of normal fibroblasts and postnatal neural progenitors, whereas downregulation of USP16 partially rescues the proliferation defects of Down’s syndrome fibroblasts. Taken together, these results suggest that USP16 has an important role in antagonizing the self-renewal and/or senescence pathways in Down’s syndrome and could serve as an attractive target to ameliorate some of the associated pathologies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Antonarakis, S. E., Lyle, R., Dermitzakis, E. T., Reymond, A. & Deutsch, S. Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nature Rev. Genet. 5, 725–738 (2004)
Yang, Q., Rasmussen, S. A. & Friedman, J. M. Mortality associated with Down’s syndrome in the USA from 1983 to 1997: a population-based study. Lancet 359, 1019–1025 (2002)
Roth, G. M., Sun, B., Greensite, F. S., Lott, I. T. & Dietrich, R. B. Premature aging in persons with Down syndrome: MR findings. Am. J. Neuroradiol. 17, 1283–1289 (1996)
Zigman, W. B. & Lott, I. T. Alzheimer’s disease in Down syndrome: neurobiology and risk. Ment. Retard. Dev. Disabil. Res. Rev. 13, 237–246 (2007)
Liu, L. & Rando, T. A. Manifestations and mechanisms of stem cell aging. J. Cell Biol. 193, 257–266 (2011)
Reeves, R. H. et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nature Genet. 11, 177–184 (1995)
Sago, H. et al. Genetic dissection of region associated with behavioral abnormalities in mouse models for Down syndrome. Pediatr. Res. 48, 606–613 (2000)
Sago, H. et al. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc. Natl Acad. Sci. USA 95, 6256–6261 (1998)
Joo, H.-Y. et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449, 1068–1072 (2007)
Lorenzo, L. P. E. et al. Defective hematopoietic stem cell and lymphoid progenitor development in the Ts65Dn mouse model of Down syndrome: potential role of oxidative stress. Antioxid. Redox Signal. 15, 2083–2094 (2011)
Chao, M. P., Seita, J. & Weissman, I. L. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb. Symp. Quant. Biol. 73, 439–449 (2008)
Akala, O. O. et al. Long-term haematopoietic reconstitution by Trp53−/−p16Ink4a−/−p19Arf−/− multipotent progenitors. Nature 453, 228–232 (2008)
Park, I.-K. et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 (2003)
Pietersen, A. M. et al. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr. Biol. 18, 1094–1099 (2008)
Molofsky, A. V. et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425, 962–967 (2003)
Cao, G. et al. Bmi-1 absence causes premature brain degeneration. PLoS ONE 7, e32015 (2012)
Lorenzi, H. A. & Reeves, R. H. Hippocampal hypocellularity in the Ts65Dn mouse originates early in development. Brain Res. 1104, 153–159 (2006)
Ellis, P. et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 26, 148–165 (2004)
Fischer, J. et al. Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nature Protocols 6, 1981–1989 (2011)
Pastrana, E., Cheng, L.-C. & Doetsch, F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc. Natl Acad. Sci. USA 106, 6387–6392 (2009)
Stingl, J. et al. Purification and unique properties of mammary epithelial stem cells. Nature 439, 993–997 (2006)
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006)
Jacobs, J. J., Kieboom, K., Marino, S., DePinho, R. A. & van Lohuizen, M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397, 164–168 (1999)
Liu, Y. et al. Expression of p16 INK4a in peripheral blood T-cells is a biomarker of human aging. Aging Cell 8, 439–448 (2009)
Krishnamurthy, J. et al. Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 114, 1299–1307 (2004)
Janzen, V. et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443, 421–426 (2006)
Kimura, M. et al. Proliferation dynamics in cultured skin fibroblasts from Down syndrome subjects. Free Radic. Biol. Med. 39, 374–380 (2005)
Carmeliet, G., David, G. & Cassiman, J. J. Cellular ageing of Alzheimer’s disease and Down syndrome cells in culture. Mutat. Res. 256, 221–231 (1991)
Arron, J. R. et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441, 595–600 (2006)
Roy, A. et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc. Natl Acad. Sci. USA 109, 17579–17584 (2012)
Reinholdt, L. G. et al. Molecular characterization of the translocation breakpoints in the Down syndrome mouse model Ts65Dn. Mamm. Genome 22, 685–691 (2011)
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009)
Dalerba, P. et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nature Biotechnol. 29, 1120–1127 (2011)
Zeng, Y. A. & Nusse, R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 6, 568–577 (2010)
Negishi, M. et al. A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS ONE 5, e12373 (2010)
Ventura, A. et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. USA 101, 10380–10385 (2004)
Shimono, Y. et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 (2009)
Tiscornia, G., Singer, O. & Verma, I. M. Production and purification of lentiviral vectors. Nature Protocols 1, 241–245 (2006)
Majeti, R., Park, C. Y. & Weissman, I. L. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 1, 635–645 (2007)
Acknowledgements
We thank S. Marro for help in establishing MEF and TTF lines; I. L. Weissman for sharing C57BL/6 CD45.1 mice; the animal core facility of SIM1, in particular A. Valledifiera and M. Alvarez. This study was supported in part by a California Institute for Regenerative Medicine (CIRM) basic biology award III, National Institutes of Health grant numbers CA100225 and CA154209, CIRM bridges fellowship (V.H.-A.), Fondazione Umberto Veronesi (B.N.d.R.), Breast Cancer Research Program sponsored by Department of Defense (S.S.), The Breast Cancer Research Foundation (BCRF), Stanford Graduate Fellowship (Y.O.) and the Down Syndrome Research and Treatment Foundation and the Fidelity Foundation (C.C.G.).
Author information
Authors and Affiliations
Contributions
M.A. and M.F.C. designed the project and wrote the manuscript. M.A. identified Usp16 in a genetic screen, constructed the lentiviral constructs and carried out bone marrow experiments. S.S. performed most of the breast experiments. S.S.M. performed most of the neural experiments. M.A., B.N.d.R., D.Q., A.K., V.H.-A., J.R. and Y.O. contributed to mouse colony handling, lentivirus preparation and mouse fibroblast experiments. M.Q. performed the imaging studies. V.M.R. provided the bone marrow specimens. S.C. and C.C.G. oversaw the neural progenitor experiments, provided animals and reagents and revised the manuscript. Project management was carried out by M.A.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
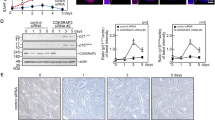
Extended Data Figure 1 Ts65Dn, but not Ts1Cje, bone marrow show a decreased number of functional HSCs.
a, Impaired engraftment ability of Ts65Dn bone marrow (BM) cells in transplantation experiments. The percentage of donor cells (CD45.2+) was evaluated at the indicated time points (five mice per group, repeated twice). b, Representative fluorescence-activated cell sorting (FACS) plots of KLS cells show a reduced number of CD34− Flt3− KLS cells in Ts65Dn marrows (four mice of each genotype were examined). c, Peripheral blood analyses four months after bone marrow transplant revealed multi-lineage engraftment in Ts65Dn mice with only 500,000 donor cells; lower doses of Ts65Dn donor bone marrow failed to reconstitute haematopoietic lineages. d, Multi-lineage analyses of peripheral blood 3 months after bone marrow transplantation showed that Ts65Dn bone marrow cells failed to reconstitute secondary recipients. Representative FACS plots are shown.
Extended Data Figure 2 HSCs in Ts65Dn mice have lower levels of H2A ubiquitination.
a, Immunofluorescence for H2A ubiquitination on Lys 119 shows a decrease in the number of positive foci in MEFs derived from Ts65Dn embryos compared to controls. Each dot represents a different cell and each column a different mouse. One hundred cells per group were analysed and the experiment was repeated twice. b, H2AK119+ staining is decreased in Ts65Dn compared to control MEFs. c, Western blot analyses of chromatin extracts from MEFs. H2AK119+ levels are decreased in Ts65Dn (quantification performed using ImageJ software). H2A western blotting verifies equal loading of extracts.
Extended Data Figure 3 Downregulation of Usp16 improves engraftment of Ts65Dn KLS cells in primary and secondary transplants.
a, Usp16 mRNA quantification after infection of KLS cells with the indicated lentivirus. b, Peripheral blood analyses revealed multi-lineage engraftment from Ts65 KLS bone marrow cells infected with a shUsp16 hairpin. Representative FACS plots are shown. c, Two months after transplantation in secondary recipients, shC Ts65Dn bone marrow cells fail to engraft, whereas shUsp16 Ts65Dn cells show multi-lineage reconstitution. Representative FACS plots are shown.
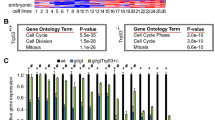
Extended Data Figure 4 Analyses of Nsp-IC frequency in neurospheres cultures.
a, Usp16 mRNA quantification in murine neurospheres cultures (P4). b, c, Raw data used for ELDA analyses of Nsp-ICs derived from Lin− SVZ cells or for the indicated sorted population. For each cell dilution, 24 replicates were tested. The table indicates the number of positive wells in each condition.
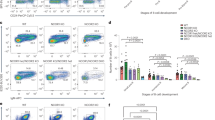
Extended Data Figure 5 CD15+ Egfr+ and Prom1+ Egfr+ populations are enriched for neuronal progenitors in mice.
a, Representative FACS plots are shown for viable Lin− cells derived from SVZ preparations. Double-positive cells were sorted and used for testing neurosphere-formation potential. b, Representative pictures of immunofluorescence staining for Sox2 and nestin on the indicated sorted populations. The arrows indicate cells scored positive for Sox2 (green) or nestin (red). For this analysis, the indicated Lin− cell populations were FACS sorted and collected by cytospin. On the right, twelve fields were randomly selected for analyses from four wild-type mice from different litters. The percentage of positive cells is given by the ratio of cells positive for Sox2 or nestin among the DAPI (4′,6-diamidino-2-phenylindole )+ cells. c, Neurosphere expansion in vitro during passaging by different sorted populations derived from mouse SVZ. CD15+ Egfr+ cells are able to expand upon passaging.
Extended Data Figure 6 Defects in mammary glands in Down’s syndrome mice models.
a, b, mRNA quantification of Usp16 and different Hox genes in CD49highCD24med mammary cells. Hox1, Hox3 and Hox5 are expressed at higher levels in Ts65Dn cells. c, Representative FACS plot of mammary cells gated on live cells (first row) or live Lin− cells. We observed a perturbation in the overall FACS profile with reduction of basal and luminal cells (indicated gates) in Ts65Dn mice but not in Ts1Cje mice. These experiments were repeated five times for each group. d, Quantification of overlap between staining for the basal cytokeratin CK14 (red) and the luminal cytokeratin CK8 (green). Pearson’s correlation analyses (lumosity software) showed a marked increase in cells that co-stain for both cytokeratins in Ts65Dn mammary epithelium. Each experiment was repeated with three mice per group. e, Downregulation of Usp16 by shRNA lentiviral infection partially rescues the in vivo defects shown by Ts65Dn mammary cells (P = 0.03). Three independent transplantation experiments were performed. Right, the percentage of fat pad filled by GFP outgrowths is significantly higher upon downregulation of Usp16 (P = 0.007).
Extended Data Figure 7 Senescence in Ts65Dn fibroblasts is affected by levels of Usp16 and Cdkn2a.
a, Western blot analyses verifies knockdown of p16. β-actin was used as a loading control. b, Proliferation of Ts65Dn TTFs increased upon infection with a hairpin targeting Cdkn2a. Control TTFs proliferate more upon downregulation of both p16Ink4a and p19Arf. The experiment was repeated three times with similar results. c, Representative pictures of p16Ink4a immunostaining (left) and quantification of the percentage of positive cells (right). Each dot represents a TTF culture derived from a different mouse. The hairpin effectively ablates p16Ink4a expression. d, SA-βgal staining in control and Ts65Dn MEFs at P4. Representative pictures are shown on the left. The percentages of positive cells are shown on the right. Experiments were replicated with three different MEF lines per genotype.
Extended Data Figure 8 Human fibroblast cultures.
a, b, mRNA quantification of USP16 upon lentiviral overexpression or downregulation. c, BMI1 overexpression significantly increases the proliferation of fibroblasts derived from a Down’s syndrome carrier. The effect on control fibroblasts is not significant. On the right, the levels of expression of BMI1 mRNA were quantified by real-time PCR. The experiment was repeated twice with similar results. d, Overexpression of USP16 reduces the formation of neurospheres derived from human paediatric SVZ cells. On the right, the quantification of the number of spheres obtained in the first and in the second passage. All the experiments were replicated twice.
Extended Data Figure 9 HSCs in human bone marrow derived from Down’s syndrome patients.
Left, representative plot of Lin− cells derived from human bone marrow. Right, a plot of the percentage of CD34+ CD38−CD90+ observed between the Lin− cells (Down’s syndrome: two samples; healthy donors: three samples). Down’s syndrome bone marrow samples were retrieved from sternal biopsies obtained from children undergoing corrective heart surgery at Stanford University. The families were consented according to IRB approved protocols. Control bone marrow samples were obtained from AllCells, LLC. Although we cannot unequivocally state that the HSCs are reduced in Down’s syndrome patients, in these children they are not increased as they might be in the liver of some human Down’s syndrome fetuses. Down’s syndrome is associated with increased rates of childhood leukaemia and decreased rates of some adult solid tumours. We can only speculate as to why Down’s syndrome predisposes individuals to one type of malignancy and protects from another, and whether USP16 has a role in either observation. Other syndromes causing bone marrow failure, such as Fanconi anaemia, predispose to leukaemia. Lymphoid leukaemias in Down’s syndrome patients frequently involve mutation of CDKN2A. Because CDKN2A appears to have a role in the proliferation defects caused by trisomy of USP16, mutations of CDKN2A could give Down’s syndrome HSCs a strong selection advantage. Differences between fetal liver and adult bone marrow HSCs could provide another explanation for an increased incidence of leukaemia in Down’s syndrome. An expansion of Down’s syndrome fetal liver HSCs but not of adult bone marrow HSCs could also explain why Down’s syndrome leukaemia appears to originate more often in fetal blood cells.
Rights and permissions
About this article
Cite this article
Adorno, M., Sikandar, S., Mitra, S. et al. Usp16 contributes to somatic stem-cell defects in Down’s syndrome. Nature 501, 380–384 (2013). https://doi.org/10.1038/nature12530
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12530
This article is cited by
-
FGF18 alleviates hepatic ischemia-reperfusion injury via the USP16-mediated KEAP1/Nrf2 signaling pathway in male mice
Nature Communications (2023)
-
MicroRNA regulation of cancer stem cells in the pathogenesis of breast cancer
Cancer Cell International (2021)
-
Restoration of keratinocytic phenotypes in autonomous trisomy-rescued cells
Stem Cell Research & Therapy (2021)
-
Deubiquitylases in developmental ubiquitin signaling and congenital diseases
Cell Death & Differentiation (2021)
-
Clinical and biological aspects of myeloid leukemia in Down syndrome
Leukemia (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.