Abstract
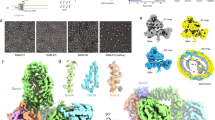
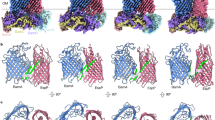
β-barrel membrane proteins are essential for nutrient import, signalling, motility and survival. In Gram-negative bacteria, the β-barrel assembly machinery (BAM) complex is responsible for the biogenesis of β-barrel membrane proteins, with homologous complexes found in mitochondria and chloroplasts. Here we describe the structure of BamA, the central and essential component of the BAM complex, from two species of bacteria: Neisseria gonorrhoeae and Haemophilus ducreyi. BamA consists of a large periplasmic domain attached to a 16-strand transmembrane β-barrel domain. Three structural features shed light on the mechanism by which BamA catalyses β-barrel assembly. First, the interior cavity is accessible in one BamA structure and conformationally closed in the other. Second, an exterior rim of the β-barrel has a distinctly narrowed hydrophobic surface, locally destabilizing the outer membrane. And third, the β-barrel can undergo lateral opening, suggesting a route from the interior cavity in BamA into the outer membrane.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dalbey, R. E., Wang, P. & Kuhn, A. Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 (2011)
White, S. H. & von Heijne, G. How translocons select transmembrane helices. Annu. Rev. Biophys. 37, 23–42 (2008)
du Plessis, D. J., Nouwen, N. & Driessen, A. J. The Sec translocase. Biochim. Biophys. Acta 1808, 851–865 (2011)
Osborne, A. R., Rapoport, T. A. & van den Berg, B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 21, 529–550 (2005)
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009)
Walther, D. M., Rapaport, D. & Tommassen, J. Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell. Mol. Life Sci. 66, 2789–2804 (2009)
Webb, C. T., Heinz, E. & Lithgow, T. Evolution of the β-barrel assembly machinery. Trends Microbiol. 20, 612–620 (2012)
Paschen, S. A., Neupert, W. & Rapaport, D. Biogenesis of β-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 30, 575–582 (2005)
Jiang, J. H., Tong, J., Tan, K. S. & Gabriel, K. From evolution to pathogenesis: the link between β-barrel assembly machineries in the outer membrane of mitochondria and Gram-negative bacteria. Int. J. Mol. Sci. 13, 8038–8050 (2012)
Tommassen, J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology 156, 2587–2596 (2010)
Pugsley, A. P. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57, 50–108 (1993)
Knowles, T. J., Scott-Tucker, A., Overduin, M. & Henderson, I. R. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nature Rev. Microbiol. 7, 206–214 (2009)
Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 (2005)
Hagan, C. L., Silhavy, T. J. & Kahne, D. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 (2011)
Ricci, D. P. & Silhavy, T. J. The Bam machine: a molecular cooper. Biochim. Biophys. Acta 1818, 1067–1084 (2012)
Rigel, N. W. & Silhavy, T. J. Making a beta-barrel: assembly of outer membrane proteins in Gram-negative bacteria. Curr. Opin. Microbiol. 15, 189–193 (2012)
Hagan, C. L., Kim, S. & Kahne, D. Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892 (2010)
Noinaj, N., Fairman, J. W. & Buchanan, S. K. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J. Mol. Biol. 407, 248–260 (2011)
Heuck, A., Schleiffer, A. & Clausen, T. Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J. Mol. Biol. 406, 659–666 (2011)
Kim, K. H., Aulakh, S. & Paetzel, M. Crystal structure of β-barrel assembly machinery BamCD protein complex. J. Biol. Chem. 286, 39116–39121 (2011)
Albrecht, R. & Zeth, K. Structural basis of outer membrane protein biogenesis in bacteria. J. Biol. Chem. 286, 27792–27803 (2011)
Sandoval, C. M., Baker, S. L., Jansen, K., Metzner, S. I. & Sousa, M. C. Crystal structure of BamD: an essential component of the β-barrel assembly machinery of gram-negative bacteria. J. Mol. Biol. 409, 348–357 (2011)
Dong, C., Hou, H. F., Yang, X., Shen, Y. Q. & Dong, Y. H. Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Crystallogr. D 68, 95–101 (2012)
Knowles, T. J. et al. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 12, 123–128 (2011)
Gatzeva-Topalova, P. Z., Walton, T. A. & Sousa, M. C. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16, 1873–1881 (2008)
Kim, S. et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 (2007)
Zhang, H. et al. High-resolution structure of a new crystal form of BamA POTRA4–5 from Escherichia coli. Acta Crystallogr. F 67, 734–738 (2011)
Gatzeva-Topalova, P. Z., Warner, L. R., Pardi, A. & Sousa, M. C. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18, 1492–1501 (2010)
Knowles, T. J. et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 68, 1216–1227 (2008)
Clantin, B. et al. Structure of the membrane protein FhaC: a member of the Omp85–TpsB transporter superfamily. Science 317, 957–961 (2007)
Fan, E., Fiedler, S., Jacob-Dubuisson, F. & Muller, M. Two-partner secretion of gram-negative bacteria: a single β-barrel protein enables transport across the outer membrane. J. Biol. Chem. 287, 2591–2599 (2012)
Rigel, N. W., Ricci, D. P. & Silhavy, T. J. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc. Natl Acad. Sci. USA 110, 5151–5156 (2013)
Leonard-Rivera, M. & Misra, R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J. Bacteriol. 194, 4662–4668 (2012)
Gumbart, J., Wiener, M. C. & Tajkhorshid, E. Coupling of calcium and substrate binding through loop alignment in the outer-membrane transporter BtuB. J. Mol. Biol. 393, 1129–1142 (2009)
Hearn, E. M., Patel, D. R., Lepore, B. W., Indic, M. & van den Berg, B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature 458, 367–370 (2009)
Cuesta-Seijo, J. A. et al. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure 18, 1210–1219 (2010)
Hong, H., Patel, D. R., Tamm, L. K. & van den Berg, B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J. Biol. Chem. 281, 7568–7577 (2006)
Tamm, L. K., Hong, H. & Liang, B. Folding and assembly of beta-barrel membrane proteins. Biochim. Biophys. Acta 1666, 250–263 (2004)
Tamm, L. K., Arora, A. & Kleinschmidt, J. H. Structure and assembly of beta-barrel membrane proteins. J. Biol. Chem. 276, 32399–32402 (2001)
Agah, S. & Faham, S. Crystallization of membrane proteins in bicelles. Methods Mol. Biol. 914, 3–16 (2012)
Ujwal, R. & Bowie, J. U. Crystallizing membrane proteins using lipidic bicelles. Methods 55, 337–341 (2011)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Stein, N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 (2008)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Ramachandran, S., Kota, P., Ding, F. & Dokholyan, N. V. Automated minimization of steric clashes in protein structures. Proteins 79, 261–270 (2011)
Shaw, D. E. et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM 51, 91–97 (2008)
Fairman, J. W. et al. Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure 20, 1233–1243 (2012)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Robert, V. et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4, e377 (2006)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Barnard, T. J. et al. Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. J. Mol. Biol. 415, 128–142 (2012)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
Mackerell, A. D., Jr, Feig, M. & Brooks, C. L., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 (2004)
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983)
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010)
Ulmschneider, J. P., Smith, J. C., White, S. H. & Ulmschneider, M. B. In silico partitioning and transmembrane insertion of hydrophobic peptides under equilibrium conditions. J. Am. Chem. Soc. 133, 15487–15495 (2011)
Elbing, K. & Brent, R. Media preparation and bacteriological tools. Curr. Protoc. Mol. Biol. Chapter 1, Unit 1.1 (2002)
Acknowledgements
We thank H. Bernstein and R. Ieva for providing JCM-166 cells, J. Beckwith and R. Misra for providing antibodies, and A. M. Stanley and T. Barnard for discussions and comments on the manuscript. N.N., A.J.K., N.C.E., H.C. and S.K.B. are supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases. J.C.G. acknowledges support from the NIH under grants K22-AI100927 and R01-GM67887. P.L. is supported by a postdoctoral fellowship through the Diamond Light Source. T.L. is an Australian Research Council (ARC) Federation Fellow and acknowledges support from ARC Discovery Project (DP120101878) and ARC Linkage International Grant (LX0776170). We thank the respective staffs at the Southeast Regional Collaborative Access Team (SER-CAT) and General Medicine and Cancer Institute’s Collaborative Access Team (GM/CA-CAT) beamlines at the Advanced Photon Source, Argonne National Laboratory and the Diamond Light Source for their assistance during data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38 (SER-CAT), and by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357 (GM/CA-CAT). Anton computer time was provided by the National Resource for Biomedical Supercomputing and the Pittsburgh Supercomputing Center through Grant RC2GM093307 from the NIH, using a machine donated by DE Shaw Research.
Author information
Authors and Affiliations
Contributions
N.N., H.C., N.C.E. and S.K.B. cloned, expressed and purified HdBamAΔ3 and NgBamA. P.L. performed data collection for experimental phasing. N.N. crystallized and solved the HdBamAΔ3 and NgBamA crystal structures. N.N. and A.J.K. performed the homology modelling and functional assays. J.C.G. designed, conducted and analysed the molecular dynamics simulations. N.N., A.J.K. and S.K.B. analysed and discussed all data. T.L. and S.K.B. conceived and designed the original project. N.N., A.J.K., T.L. and S.K.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2, Supplementary Figures 1-18, Supplementary Models 1-2, details of Supplementary Videos 1-2 and additional references. (PDF 4312 kb)
Supplementary Data
This file contains the data for Supplementary Models 1-2. (ZIP 169 kb)
Molecular dynamics (MD) simulations of FhaC and NgBamA
MD simulations were used to probe the stability of the barrel domains of FhaC and NgBamA. This video illustrates the changes that occur within the barrel domain (opening and closing) in NgBamA as the simulation progresses (see also Figure 4 and Supplementary Figure 16). (MOV 10499 kb)
Proposed mechanisms for the role of BamA in the biogenesis of β-barrel membrane proteins.
Animated video summarizing the two proposed mechanisms believed to be involved in recognizing, folding, and inserting β-barrel membrane proteins into the outer membrane. These mechanisms are based on studies (structural biology, computational, and functional) reported here, as well as, previously reported studies about the function of BamA. (MOV 5810 kb)
Rights and permissions
About this article
Cite this article
Noinaj, N., Kuszak, A., Gumbart, J. et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 (2013). https://doi.org/10.1038/nature12521
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12521
This article is cited by
-
Surveying membrane landscapes: a new look at the bacterial cell surface
Nature Reviews Microbiology (2023)
-
Mechanisms of membrane protein crystallization in ‘bicelles’
Scientific Reports (2022)
-
Rhamnolipid the Glycolipid Biosurfactant: Emerging trends and promising strategies in the field of biotechnology and biomedicine
Microbial Cell Factories (2021)
-
Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily
Nature Structural & Molecular Biology (2021)
-
The role of membrane destabilisation and protein dynamics in BAM catalysed OMP folding
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.