Abstract
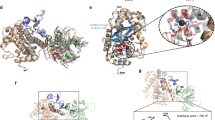
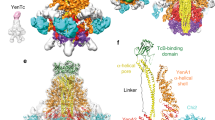
The ABC toxin complexes produced by certain bacteria are of interest owing to their potent insecticidal activity1,2 and potential role in human disease3. These complexes comprise at least three proteins (A, B and C), which must assemble to be fully toxic4. The carboxy-terminal region of the C protein is the main cytotoxic component5, and is poorly conserved between different toxin complexes. A general model of action has been proposed, in which the toxin complex binds to the cell surface via the A protein, is endocytosed, and subsequently forms a pH-triggered channel, allowing the translocation of C into the cytoplasm, where it can cause cytoskeletal disruption in both insect and mammalian cells5. Toxin complexes have been visualized using single-particle electron microscopy6,7, but no high-resolution structures of the components are available, and the role of the B protein in the mechanism of toxicity remains unknown. Here we report the three-dimensional structure of the complex formed between the B and C proteins, determined to 2.5 Å by X-ray crystallography. These proteins assemble to form an unprecedented, large hollow structure that encapsulates and sequesters the cytotoxic, C-terminal region of the C protein like the shell of an egg. The shell is decorated on one end by a β-propeller domain, which mediates attachment of the B–C heterodimer to the A protein in the native complex. The structure reveals how C auto-proteolyses when folded in complex with B. The C protein is the first example, to our knowledge, of a structure that contains rearrangement hotspot (RHS) repeats8, and illustrates a marked structural architecture that is probably conserved across both this widely distributed bacterial protein family and the related eukaryotic tyrosine-aspartate (YD)-repeat-containing protein family, which includes the teneurins9. The structure provides the first clues about the function of these protein repeat families, and suggests a generic mechanism for protein encapsulation and delivery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Accessions
Protein Data Bank
Data deposits
The atomic coordinates of the B–CNTR complex and the Chi2 chitinase have been deposited in the Protein Data Bank (http://www.pdb.org) under the accession codes 4IGL and 4DWS, respectively.
References
Bowen, D. et al. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280, 2129–2132 (1998)
ffrench-Constant, R. H. & Waterfield, N. R. Ground control for insect pests. Nature Biotechnol. 24, 660–661 (2006)
Pinheiro, V. B. & Ellar, D. J. Expression and insecticidal activity of Yersinia pseudotuberculosis and Photorhabdus luminescens toxin complex proteins. Cell. Microbiol. 9, 2372–2380 (2007)
Waterfield, N., Hares, M., Yang, G., Dowling, A. & ffrench-Constant, R. Potentiation and cellular phenotypes of the insecticidal toxin complexes of Photorhabdus bacteria. Cell. Microbiol. 7, 373–382 (2005)
Lang, A. E. et al. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science 327, 1139–1142 (2010)
Landsberg, M. J. et al. 3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity. Proc. Natl Acad. Sci. USA 108, 20544–20549 (2011)
Gatsogiannis, C. et al. A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature 495, 520–523 (2013)
Hill, C. W., Sandt, C. H. & Vlazny, D. A. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12, 865–871 (1994)
Minet, A. D., Rubin, B. P., Tucker, R. P., Baumgartner, S. & Chiquet-Ehrismann, R. Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. J. Cell Sci. 112, 2019–2032 (1999)
Hurst, M. R. H. et al. The main virulence determinant of Yersinia entomophaga MH96 is a broad-host-range toxin complex active against insects. J. Bacteriol. 193, 1966–1980 (2011)
Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M. & Aravind, L. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 (2012)
Buetow, L., Flatau, G., Chiu, K., Boquet, P. & Ghosh, P. Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1. Nature Struct. Biol. 8, 584–588 (2001)
Iyer, L. M., Zhang, D., Rogozin, I. B. & Aravind, L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 39, 9473–9497 (2011)
Jackson, A. P., Thomas, G. H., Parkhill, J. & Thomson, N. R. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics 10, 584 (2009)
Wang, Y. D., Zhao, S. & Hill, C. W. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J. Bacteriol. 180, 4102–4110 (1998)
Mosca, T. J., Hong, W., Dani, V. S., Favaloro, V. & Luo, L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 484, 237–241 (2012)
Hong, W., Mosca, T. J. & Luo, L. Teneurins instruct synaptic partner matching in an olfactory map. Nature 484, 201–207 (2012)
Feng, K. et al. All four members of the Ten-m/Odz family of transmembrane proteins form dimers. J. Biol. Chem. 277, 26128–26135 (2002)
Chand, D. et al. C-terminal processing of the teneurin proteins: Independent actions of a teneurin C-terminal associated peptide in hippocampal cells. Mol. Cell. Neurosci. 52, 38–50 (2013)
Tucker, R. P. & Chiquet-Ehrismann, R. Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev. Biol. 290, 237–245 (2006)
Busby, J. N. et al. Structural analysis of Chi1 chitinase from Yen-Tc: the multisubunit insecticidal ABC toxin complex of Yersinia entomophaga. J. Mol. Biol. 415, 359–371 (2012)
Xu, Z., Horwich, A. L. & Sigler, P. B. The crystal structure of the asymmetric GroEL–GroES–(ADP)7 chaperonin complex. Nature 388, 741–750 (1997)
McPhillips, T. M. et al. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 (2002)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
The. CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Panjikar, S., Parthasarathy, V., Lamzin, V. S., Weiss, M. S. & Tucker, P. A. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D 61, 449–457 (2005)
Panjikar, S., Parthasarathy, V., Lamzin, V. S., Weiss, M. S. & Tucker, P. A. On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr. D 65, 1089–1097 (2009)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Orthaber, D., Bergmann, A. & Glatter, O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Crystallogr. 33, 218–225 (2000)
Franke, D. & Svergun, D. I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009)
Volkov, V. V. & Svergun, D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003)
Svergun, D. I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999)
Svergun, D., Barberato, C. & Koch, M. H. J. CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995)
Scheres, S. H. W., Nuñez-Ramirez, R., Sorzano, C. O. S., Carazo, J. M. & Marabini, R. Image processing for electron microscopy single-particle analysis using Xmipp. Nature Protocols 3, 977–990 (2008)
Acknowledgements
We are grateful to M. Sullivan, R. Kingston, T. Baker and many other members of the Structural Biology section at the University of Auckland for discussions, to V. Arcus for initial enthusiasm, and to B. Hankamer for provision of laboratory space at the University of Queensland and for mentorship. This work was supported by the New Zealand Foundation for Research, Science and Technology contract C10X0804, awarded to M.R.H.H. We would like to thank all beamline staff at the MX1, MX2 and SAXS/WAXS beamlines at the Australian Synchrotron for their support, and to the New Zealand Synchrotron Group Ltd for synchrotron access arrangements.
Author information
Authors and Affiliations
Contributions
J.N.B. cloned constructs, expressed, purified and crystallized proteins, collected and processed X-ray crystallography and SAXS data, and refined and analysed the protein structure; S.P. determined the protein structure; M.J.L. processed, refined and analysed negative-stain electron microscopy data; M.R.H.H. and J.S.L. designed the study; J.N.B. and J.S.L. wrote the paper and all authors discussed the results and contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-5, Supplementary Figures 1-12 and additional references. (PDF 5176 kb)
Rights and permissions
About this article
Cite this article
Busby, J., Panjikar, S., Landsberg, M. et al. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 501, 547–550 (2013). https://doi.org/10.1038/nature12465
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12465
This article is cited by
-
Stepwise assembly and release of Tc toxins from Yersinia entomophaga
Nature Microbiology (2024)
-
CRISPR screens in Drosophila cells identify Vsg as a Tc toxin receptor
Nature (2022)
-
The P. aeruginosa effector Tse5 forms membrane pores disrupting the membrane potential of intoxicated bacteria
Communications Biology (2022)
-
Pseudomonas fluorescens F113 type VI secretion systems mediate bacterial killing and adaption to the rhizosphere microbiome
Scientific Reports (2021)
-
Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.