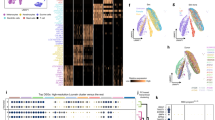
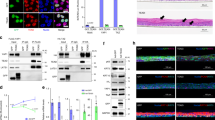
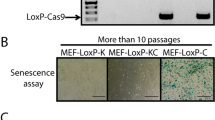
Abstract
Tissue growth is the multifaceted outcome of a cell’s intrinsic capabilities and its interactions with the surrounding environment. Decoding these complexities is essential for understanding human development and tumorigenesis. Here we tackle this problem by carrying out the first genome-wide RNA-interference-mediated screens in mice. Focusing on skin development and oncogenic (HrasG12V-induced) hyperplasia, our screens uncover previously unknown as well as anticipated regulators of embryonic epidermal growth. Among the top oncogenic screen hits are Mllt6 and the Wnt effector β-catenin, which maintain HrasG12V-dependent hyperproliferation. We also expose β-catenin as an unanticipated antagonist of normal epidermal growth, functioning through Wnt-independent intercellular adhesion. Finally, we validate functional significance in mouse and human cancers, thereby establishing the feasibility of in vivo mammalian genome-wide investigations to dissect tissue development and tumorigenesis. By documenting some oncogenic growth regulators, we pave the way for future investigations of other hits and raise promise for unearthing new targets for cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
Gene Expression Omnibus
Data deposits
Raw RNAseq data can be accessed at Gene Expression Omnibus under accession number GSE48480 and permissions information is available at http://www.nature.com/reprints.
References
Beronja, S., Livshits, G., Williams, S. & Fuchs, E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nature Med. 16, 821–827 (2010)
Williams, S. E., Beronja, S., Pasolli, H. A. & Fuchs, E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353–358 (2011)
Ezratty, E. J. et al. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell 145, 1129–1141 (2011)
Oshimori, N. & Fuchs, E. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 10, 63–75 (2012)
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001)
Karnoub, A. E. & Weinberg, R. A. Ras oncogenes: split personalities. Nature Rev. Mol. Cell Biol. 9, 517–531 (2008)
Balmain, A. & Pragnell, I. B. Mouse skin carcinomas induced in vivo by chemical carcinogens have a transforming Harvey-ras oncogene. Nature 303, 72–74 (1983)
Vasioukhin, V., Degenstein, L., Wise, B. & Fuchs, E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl Acad. Sci. USA 96, 8551–8556 (1999)
Chen, X. et al. Endogenous expression of HrasG12V induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc. Natl Acad. Sci. USA 106, 7979–7984 (2009)
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997)
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006)
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010)
Donninger, H. et al. The Ras effector RASSF2 controls the PAR-4 tumor suppressor. Mol. Cell. Biol. 30, 2608–2620 (2010)
Miyata, Y. et al. Cyclin C regulates human hematopoietic stem/progenitor cell quiescence. Stem Cells 28, 308–317 (2010)
Prasad, R. et al. Leucine-zipper dimerization motif encoded by the AF17 gene fused to ALL-1 (MLL) in acute leukemia. Proc. Natl Acad. Sci. USA 91, 8107–8111 (1994)
Gat, U., DasGupta, R., Degenstein, L. & Fuchs, E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell 95, 605–614 (1998)
Chan, E. F., Gat, U., McNiff, J. M. & Fuchs, E. A common human skin tumour is caused by activating mutations in beta-catenin. Nature Genet. 21, 410–413 (1999)
Malanchi, I. et al. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature 452, 650–653 (2008)
Nusse, R. Wnt signaling and stem cell control. Cell Res. 18, 523–527 (2008)
Huelsken, J., Vogel, R., Erdmann, B., Cotsarelis, G. & Birchmeier, W. β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 105, 533–545 (2001)
Nguyen, H. et al. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nature Genet. 41, 1068–1075 (2009)
Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012)
Posthaus, H. et al. β-Catenin is not required for proliferation and differentiation of epidermal mouse keratinocytes. J. Cell Sci. 115, 4587–4595 (2002)
Vasioukhin, V., Bauer, C., Yin, M. & Fuchs, E. Direct actin polymerization is the driving force for epithelial cell–cell adhesion. Cell 100, 209–219 (2000)
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signaling. Nature 461, 614–620 (2009)
DasGupta, R. & Fuchs, E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568 (1999)
Kandyba, E. et al. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc. Natl Acad. Sci. USA 110, 1351–1356 (2013)
Schober, M. & Fuchs, E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-β and integrin/focal adhesion kinase (FAK) signaling. Proc. Natl Acad. Sci. USA 108, 10544–10549 (2011)
Scholl, C. et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821–834 (2009)
Babij, C. et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 71, 5818–5826 (2011)
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011)
Wang, Y. et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science 327, 1650–1653 (2010)
Suvà, M. L., Riggi, N. & Bernstein, B. E. Epigenetic reprograming in cancer. Science 339, 1567–1570 (2013)
Bernt, K. M. et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011)
Beronja, S. & Fuchs, E. RNAi-mediated gene function analysis in skin. Methods Mol. Biol. 961, 351––361 (2013)
Linkert, M. et al. Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010)
Nowak, J. A. & Fuchs, E. Isolation and culture of epithelial stem cells. Methods Mol. Biol. 482, 215–232 (2009)
Wiederschain, D. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009)
Davidson, K. C. et al. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl Acad. Sci. USA 109, 4485–4490 (2012)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Auer, P. L. & Doerge, R. W. Statistical design and analysis of RNA sequencing data. Genetics 185, 405–416 (2010)
R Research Development Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org (2012)
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009)
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 28, 511–515 (2010)
Acknowledgements
We thank J. Fagin for inducible oncogenic Hras mice; S. Williams, M. Schober, A. Rodriguez Folgueras, S. Dewell and D. Schramek for intellectual input; D. Oristian and N. Stokes as mouse specialists; Comparative Bioscience Center (AAALAC accredited) for care of mice in accordance with National Institutes of Health (NIH) guidelines; Genomics Resource Center (C. Zhao, Director) for sequencing; Bioimaging Center (A. North, Director) for advice; Flow Cytometry facility (S. Mazel, Director) for FACS sorting. E.F. is an Investigator of the Howard Hughes Medical Institute. This research was supported by grants from the NIH (R37-AR27883 (E.F.) and K99-AR061469 (S.B.)), Emerald Foundation (E.F.) and Human Frontiers Science Program Postdoctoral Fellowship (S.B.).
Author information
Authors and Affiliations
Contributions
S.B., P.J. and E.F. designed the experiments. S.B. and P.J. made shRNA pools and lentivirus, and performed the screens. Illumina sequence analysis was done by E.H. and S.B. RNAseq and IPA analyses were performed by S.B., B.E.K. and P.J. Imaging was done by S.B. and N.O., and image analysis by S.B. and E.H. CHIP-seq data was generated by W.-H.L., and P.J. performed luciferase assays. S.B. and E.F. wrote the paper. All authors provided intellectual input, vetted and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-20. (PDF 27465 kb)
Supplementary Tables
This file contains Supplementary Tables 1-5. The page numbers for each of the tables are as follows: 1) 1-71, 2) 72-180, 3) 181-188, 4) 189-200 and 5) 201. (PDF 6474 kb)
Supplementary Data
This file contains searchable lists of genes presented in Supplementary Tables 1-4. (XLSX 554 kb)
Rights and permissions
About this article
Cite this article
Beronja, S., Janki, P., Heller, E. et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501, 185–190 (2013). https://doi.org/10.1038/nature12464
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12464
This article is cited by
-
The role of noncoding RNAs in metabolic reprogramming of cancer cells
Cellular & Molecular Biology Letters (2023)
-
In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer
Nature Communications (2023)
-
ROR2 regulates self-renewal and maintenance of hair follicle stem cells
Nature Communications (2022)
-
Mechanics of a multilayer epithelium instruct tumour architecture and function
Nature (2020)
-
RNaseH-mediated simultaneous piggyback knockdown of multiple genes in adult zebrafish
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.