Abstract
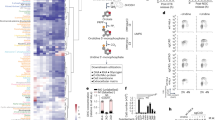
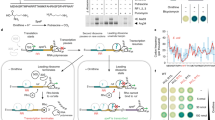
Cellular metabolism converts available nutrients into usable energy and biomass precursors. The process is regulated to facilitate efficient nutrient use and metabolic homeostasis. Feedback inhibition of the first committed step of a pathway by its final product is a classical means of controlling biosynthesis1,2,3,4. In a canonical example, the first committed enzyme in the pyrimidine pathway in Escherichia coli is allosterically inhibited by cytidine triphosphate1,4,5. The physiological consequences of disrupting this regulation, however, have not been previously explored. Here we identify an alternative regulatory strategy that enables precise control of pyrimidine pathway end-product levels, even in the presence of dysregulated biosynthetic flux. The mechanism involves cooperative feedback regulation of the near-terminal pathway enzyme uridine monophosphate kinase6. Such feedback leads to build-up of the pathway intermediate uridine monophosphate, which is in turn degraded by a conserved phosphatase, here termed UmpH, with previously unknown physiological function7,8. Such directed overflow metabolism allows homeostasis of uridine triphosphate and cytidine triphosphate levels at the expense of uracil excretion and slower growth during energy limitation. Disruption of the directed overflow regulatory mechanism impairs growth in pyrimidine-rich environments. Thus, pyrimidine homeostasis involves dual regulatory strategies, with classical feedback inhibition enhancing metabolic efficiency and directed overflow metabolism ensuring end-product homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gerhart, J. C. & Pardee, A. B. The enzymology of control by feedback inhibition. J. Biol. Chem. 237, 891–896 (1962)
Savageau, M. A. Optimal design of feedback-control by inhibition — dynamic considerations. J. Mol. Evol. 5, 199–222 (1975)
Umbarger, H. E. Evidence for a negative-feedback mechanism in the biosynthesis of isoleucine. Science 123, 848–849 (1956)
Pardee, A. B. & Yates, R. A. Control of pyrimidine biosynthesis in Escherichia coli by a feed-back mechanism. J. Biol. Chem. 221, 757–770 (1956)
Kantrowitz, E. R. Allostery and cooperativity in Escherichia coli aspartate transcarbamoylase. Arch. Biochem. Biophys. 519, 81–90 (2012)
Meyer, P. et al. Structural and functional characterization of Escherichia coli UMP kinase in complex with its allosteric regulator GTP. J. Biol. Chem. 283, 36011–36018 (2008)
Kuznetsova, E. et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 281, 36149–36161 (2006)
Proudfoot, M. et al. General enzymatic screens identify three new nucleotidases in Escherichia coli. Biochemical characterization of SurE, YfbR, and YjjG. J. Biol. Chem. 279, 54687–54694 (2004)
Orth, J. D. et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 7, 535 (2011)
Fell, D. Understanding the Control of Metabolism (Portland Press, 1997)
Kacser, H., Burns, J. A. & Fell, D. A. The control of flux. Biochem. Soc. Trans. 23, 341–366 (1995)
Heinrich, R. & Rapoport, T. A. Linear steady-state treatment of enzymatic chains. General properties, control and effector strength. Eur. J. Biochem. 42, 89–95 (1974)
Crabtree, B. & Newsholme, E. A. The derivation and interpretation of control coefficients. Biochem. J. 247, 113–120 (1987)
Small, J. R. & Kacser, H. Responses of metabolic systems to large changes in enzyme-activities and effectors. 1. The linear treatment of unbranched chains. Eur. J. Biochem. 213, 613–624 (1993)
Kell, D. B. & Westerhoff, H. V. Metabolic control theory: its role in microbiology and biotechnology. FEMS Microbiol. Rev. 39, 305–320 (1986)
Hofmeyr, J. H. S. & Cornish-Bowden, A. Quantitative assessment of regulation in metabolic systems. Eur. J. Biochem. 200, 223–236 (1991)
Keseler, I. M. et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39, D583–D590 (2011)
Peterson, A. W., Cockrell, G. M. & Kantrowitz, E. R. A second allosteric site in Escherichia coli aspartate transcarbamoylase. Biochemistry 51, 4776–4778 (2012)
Wild, J. R., Loughrey-Chen, S. J. & Corder, T. S. In the presence of CTP, UTP becomes an allosteric inhibitor of aspartate transcarbamoylase. Proc. Natl Acad. Sci. USA 86, 46–50 (1989)
Anderson, P. M. & Meister, A. Control of Escherichia coli carbamyl phosphate synthetase by purine and pyrimidine nucleotides. Biochemistry 5, 3164–3169 (1966)
Delannay, S. et al. Serine 948 and threonine 1042 are crucial residues for allosteric regulation of Escherichia coli carbamoylphosphate synthetase and illustrate coupling effects of activation and inhibition pathways. J. Mol. Biol. 286, 1217–1228 (1999)
Loh, K. D. et al. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl Acad. Sci. USA 103, 5114–5119 (2006)
Bennett, B. D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature Chem. Biol. 5, 593–599 (2009)
Flamholz, A., Noor, E., Bar-Even, A. & Milo, R. eQuilibrator–the biochemical thermodynamics calculator. Nucleic Acids Res. 40, D770–D775 (2012)
Bianchi, V. & Spychala, J. Mammalian 5′-nucleotidases. J. Biol. Chem. 278, 46195–46198 (2003)
Tremblay, L. W., Dunaway-Mariano, D. & Allen, K. N. Structure and activity analyses of Escherichia coli K-12 NagD provide insight into the evolution of biochemical function in the haloalkanoic acid dehalogenase superfamily. Biochemistry 45, 1183–1193 (2006)
Bucurenci, N. et al. Mutational analysis of UMP kinase from Escherichia coli. J. Bacteriol. 180, 473–477 (1998)
Sauer, U. & Eikmanns, B. J. The PEP–pyruvate–oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29, 765–794 (2005)
Roche, T. E. & Hiromasa, Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell. Mol. Life Sci. 64, 830–849 (2007)
Soupene, E. et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185, 5611–5626 (2003)
Silhavy, T. J., Berman, M. L. & Enquist, L. W. Experiments With Gene Fusions (Cold Spring Harbor Press, 1984)
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006)
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000)
Kitagawa, M. et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete Set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299 (2006)
Gutnick, D., Calvo, J. M., Klopotowski, T. & Ames, B. N. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100, 215–219 (1969)
Neidhardt, F. C., Bloch, P. L. & Smith, D. F. Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974)
Rabinowitz, J. D. & Kimball, E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79, 6167–6173 (2007)
Bennett, B. D., Yuan, J., Kimball, E. H. & Rabinowitz, J. D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nature Protocols 3, 1299–1311 (2008)
Lu, W. et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 82, 3212–3221 (2010)
Lu, W., Kimball, E. & Rabinowitz, J. D. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. J. Am. Soc. Mass Spectrom. 17, 37–50 (2006)
Clasquin, M. F., Melamud, E. & Rabinowitz, J. D. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics 14, 14.11 (2012)
Goodarzi, H. et al. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol. Syst. Biol. 6, 378 (2010)
Goodarzi, H., Elemento, O. & Tavazoie, S. Revealing global regulatory perturbations across human cancers. Mol. Cell 36, 900–911 (2009)
Acknowledgements
M.L.R. was supported by an National Science foundation (NSF) Graduate Research Fellowship. J.D.R. was supported by NSF CDI Award CBET-0941143 and CAREER Award MCB-0643859, and DOE-AFOSR Award DE-SC0002077/FA9550-09-1-0580, and an American Heart Association Scientist Development Grant.
Author information
Authors and Affiliations
Contributions
M.L.R., A.M.H., B.D.Y., Y.-F.X. and J.D.R. designed experiments and analyses. B.D.Y. generated feedback-dysregulated strains and performed experiments on regulation of the de novo pathway. A.M.H. performed competitions, microarrays and metabolite quantification. M.L.R. generated overflow and cooperativity mutants and measured their metabolites and growth. M.L.R. and J.D.R. wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-6. (PDF 2376 kb)
Rights and permissions
About this article
Cite this article
Reaves, M., Young, B., Hosios, A. et al. Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature 500, 237–241 (2013). https://doi.org/10.1038/nature12445
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12445
This article is cited by
-
Cross-feeding promotes heterogeneity within yeast cell populations
Nature Communications (2024)
-
Insight into de-regulation of amino acid feedback inhibition: a focus on structure analysis method
Microbial Cell Factories (2023)
-
Metabolome and proteome analyses reveal transcriptional misregulation in glycolysis of engineered E. coli
Nature Communications (2021)
-
Temporal evolution of master regulator Crp identifies pyrimidines as catabolite modulator factors
Nature Communications (2021)
-
Microbial 5′-nucleotidases: their characteristics, roles in cellular metabolism, and possible practical applications
Applied Microbiology and Biotechnology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.