Abstract
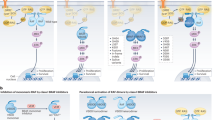
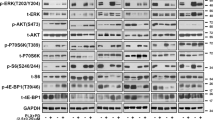
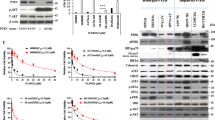
KRAS and BRAF activating mutations drive tumorigenesis through constitutive activation of the MAPK pathway. As these tumours represent an area of high unmet medical need, multiple allosteric MEK inhibitors, which inhibit MAPK signalling in both genotypes, are being tested in clinical trials. Impressive single-agent activity in BRAF-mutant melanoma has been observed; however, efficacy has been far less robust in KRAS-mutant disease1. Here we show that, owing to distinct mechanisms regulating MEK activation in KRAS- versus BRAF-driven tumours2,3, different mechanisms of inhibition are required for optimal antitumour activity in each genotype. Structural and functional analysis illustrates that MEK inhibitors with superior efficacy in KRAS-driven tumours (GDC-0623 and G-573, the former currently in phase I clinical trials) form a strong hydrogen-bond interaction with S212 in MEK that is critical for blocking MEK feedback phosphorylation by wild-type RAF. Conversely, potent inhibition of active, phosphorylated MEK is required for strong inhibition of the MAPK pathway in BRAF-mutant tumours, resulting in superior efficacy in this genotype with GDC-0973 (also known as cobimetinib), a MEK inhibitor currently in phase III clinical trials. Our study highlights that differences in the activation state of MEK in KRAS-mutant tumours versus BRAF-mutant tumours can be exploited through the design of inhibitors that uniquely target these distinct activation states of MEK. These inhibitors are currently being evaluated in clinical trials to determine whether improvements in therapeutic index within KRAS versus BRAF preclinical models translate to improved clinical responses in patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Akinleye, A., Furqan, M., Mukhi, N., Ravella, P. & Liu, D. MEK and the inhibitors: from bench to bedside. J. Hematol. Oncol. 6, 27 (2013)
Blasco, R. B. et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663 (2011)
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002)
Downward, J. Targeting RAS signalling pathways in cancer therapy. Nature Rev. Cancer 3, 11–22 (2003)
Bamford, S. et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 91, 355–358 (2004)
Hatzivassiliou, G. et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 (2010)
Heidorn, S. J. et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140, 209–221 (2010)
Poulikakos, P. I., Zhang, C., Bollag, G., Shokat, K. M. & Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010)
Solit, D. B. et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature 439, 358–362 (2006)
Wellbrock, C., Karasarides, M. & Marais, R. The RAF proteins take centre stage. Nature Rev. Mol. Cell Biol. 5, 875–885 (2004)
Chapman, M. S. & Miner, J. N. Novel mitogen-activated protein kinase kinase inhibitors. Expert Opin. Investig. Drugs 20, 209–220 (2011)
Engelman, J. A. et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nature Med. 14, 1351–1356 (2008)
Gilmartin, A. G. et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 17, 989–1000 (2011)
Hoeflich, K. P. et al. Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer Res. 72, 210–219 (2012)
Friday, B. B. et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 68, 6145–6153 (2008)
Pratilas, C. A. et al. V600EBRAF is associated with disabled feedback inhibition of RAF–MEK signaling and elevated transcriptional output of the pathway. Proc. Natl Acad. Sci. USA 106, 4519–4524 (2009)
Delaney, A. M., Printen, J. A., Chen, H., Fauman, E. B. & Dudley, D. T. Identification of a novel mitogen-activated protein kinase kinase activation domain recognized by the inhibitor PD 184352. Mol. Cell. Biol. 22, 7593–7602 (2002)
Heald, R. A. et al. Discovery of novel allosteric mitogen-activated protein kinase kinase (MEK) 1,2 inhibitors possessing bidentate Ser212 interactions. J. Med. Chem. 55, 4594–4604 (2012)
O'Hagan, D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem. Soc. Rev. 37, 308–319 (2008)
Gopalbhai, K. et al. Negative regulation of MAPKK by phosphorylation of a conserved serine residue equivalent to Ser212 of MEK1. J. Biol. Chem. 278, 8118–8125 (2003)
Sheth, P. R. et al. Fully activated MEK1 exhibits compromised affinity for binding of allosteric inhibitors U0126 and PD0325901. Biochemistry 50, 7964–7976 (2011)
Resing, K. A. & Ahn, N. G. Deuterium exchange mass spectrometry as a probe of protein kinase activation. Analysis of wild-type and constitutively active mutants of MAP kinase kinase-1. Biochemistry 37, 463–475 (1998)
Rice, K. D. et al. Novel carboxamide-based allosteric MEK inhibitors: discovery and optimization efforts toward XL518 (GDC-0973). ACS Med. Chem. Lett. 3, 416–421 (2012)
Alessi, D. R., Cuenda, A., Cohen, P., Dudley, D. T. & Saltiel, A. R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270, 27489–27494 (1995)
Ohren, J. F. et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nature Struct. Mol. Biol. 11, 1192–1197 (2004)
Garnett, M. J., Rana, S., Paterson, H., Barford, D. & Marais, R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell 20, 963–969 (2005)
Röring, M. et al. Distinct requirement for an intact dimer interface in wild-type, V600E and kinase-dead B-Raf signalling. EMBO J. 31, 2629–2647 (2012)
Rosen, L. et al. A first-in-human phase 1 study to evaluate the MEK1/2 inhibitor GDC-0973 administered daily in patients with advanced solid tumors. Cancer Res. 71, 4716 (2011)
Choo, E. F. et al. Preclinical disposition and pharmacokinetics-pharmacodynamic modeling of biomarker response and tumour growth inhibition in xenograft mouse models of G-573, a MEK inhibitor. Xenobiotica 40, 751–762 (2010)
Acknowledgements
The authors would like to acknowledge the Genentech IVCC group, IVP dosing group, as well as B. Liu and I. Yen and for technical support. SSRL is supported by the Department of Energy, the National Institutes of Health, the National Institute of General Medical Sciences and the National Center for Research Resources.
Author information
Authors and Affiliations
Contributions
J.R.H., K.S., S-M.L., J.C., A.P., C.O. J.F.M.H., D.J.A., M.J.C.L., M.Z. and M.U. performed the experiments; M.M., S.P., R.H., C.W., M.Z. and K.P.H. analysed data; L.S.F. provided comments; G.H., J.R.H., H.C., S.M. and M.B. directed the studies, analysed data, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors were all employees of Genentech or Argenta at the time that the work was carried out.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1-13 and Supplementary Tables 1-3. This file was replaced on September 11, as Supplementary Figure 7 was corrupted and Supplementary Figures 12 and 13 were missing from the original file posted on line. (PDF 10886 kb)
Rights and permissions
About this article
Cite this article
Hatzivassiliou, G., Haling, J., Chen, H. et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature 501, 232–236 (2013). https://doi.org/10.1038/nature12441
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12441
This article is cited by
-
Live-cell target engagement of allosteric MEKi on MEK–RAF/KSR–14-3-3 complexes
Nature Chemical Biology (2024)
-
Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies
Signal Transduction and Targeted Therapy (2023)
-
Low-level laser selectively inhibiting colorectal cancer cell metabolic activity and inducing apoptosis for delaying the development of intestinal cancer
Photochemical & Photobiological Sciences (2023)
-
A landscape of response to drug combinations in non-small cell lung cancer
Nature Communications (2023)
-
In silico high throughput screening and in vitro validation of a novel Raf/Mek dual inhibitor against colorectal carcinoma
Biologia (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.