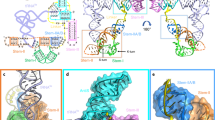
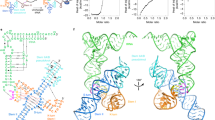
Abstract
In Gram-positive bacteria, T-box riboswitches regulate the expression of aminoacyl-tRNA synthetases and other proteins in response to fluctuating transfer RNA aminoacylation levels under various nutritional states1. T-boxes reside in the 5′-untranslated regions of the messenger RNAs they regulate, and consist of two conserved domains. Stem I contains the specifier trinucleotide that base pairs with the anticodon of cognate tRNA. 3′ to stem I is the antiterminator domain, which base pairs with the tRNA acceptor end and evaluates its aminoacylation state2. Despite high phylogenetic conservation and widespread occurrence in pathogens, the structural basis of tRNA recognition3,4 by this riboswitch remains ill defined. Here we demonstrate that the ∼100-nucleotide T-box stem I is necessary and sufficient for specific, high-affinity (dissociation constant (Kd) ∼150 nM) tRNA binding, and report the structure of Oceanobacillus iheyensis glyQ stem I in complex with its cognate tRNA at 3.2 Å resolution. Stem I recognizes the overall architecture of tRNA in addition to its anticodon, something accomplished by large ribonucleoproteins such as the ribosome, or proteins such as aminoacyl-tRNA synthetases5, but is unprecedented for a compact mRNA domain. The C-shaped stem I cradles the L-shaped tRNA, forming an extended (1,604 Å2) intermolecular interface. In addition to the specifier–anticodon interaction, two interdigitated T-loops near the apex of stem I stack on the tRNA elbow in a manner analogous to those of the J11/12–J12/11 motif6 of RNase P and the L1 stalk7 of the ribosomal E-site. Because these ribonucleoproteins and T-boxes are unrelated, this strategy to recognize a universal tRNA feature probably evolved convergently. Mutually induced fit of stem I and the tRNA exploiting the intrinsic flexibility of tRNA and its conserved post-transcriptional modifications results in high shape complementarity, which in addition to providing specificity and affinity, globally organizes the T-box to orchestrate tRNA-dependent transcription regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Green, N. J., Grundy, F. J. & Henkin, T. M. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 584, 318–324 (2010)
Grundy, F. J. & Henkin, T. M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74, 475–482 (1993)
Yousef, M. R., Grundy, F. J. & Henkin, T. M. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 349, 273–287 (2005)
Yousef, M. R., Grundy, F. J. & Henkin, T. M. tRNA requirements for glyQS antitermination: a new twist on tRNA. RNA 9, 1148–1156 (2003)
Perona, J. J. & Hadd, A. Structural diversity and protein engineering of the aminoacyl-tRNA synthetases. Biochemistry 51, 8705–8729 (2012)
Reiter, N. J. et al. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468, 784–789 (2010)
Korostelev, A., Trakhanov, S., Laurberg, M. & Noller, H. Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077 (2006)
Winkler, W. C., Grundy, F. J., Murphy, B. A. & Henkin, T. M. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7, 1165–1172 (2001)
Winkler, W. RNA Elements Required for T Box Antitermination. PhD thesis, Ohio State Univ. (2002)
Lehmann, J., Jossinet, F. & Gautheret, D. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNase P and T-box leaders. Nucleic Acids Res. 41, 5494–5502 (2013)
Gutiérrez-Preciado, A., Henkin, T. M., Grundy, F. J., Yanofsky, C. & Merino, E. Biochemical features and functional implications of the RNA-based T-Box regulatory mechanism. Microbiol. Mol. Biol. Rev. 73, 36–61 (2009)
Klein, D. J., Schmeing, T. M., Moore, P. B. & Steitz, T. A. The kink-turn: a new RNA secondary structure motif. EMBO J. 20, 4214–4221 (2001)
Baird, N. J., Zhang, J., Hamma, T. & Ferré-D’Amaré, A. R. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA 18, 759–770 (2012)
Leontis, N. B. & Westhof, E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA 4, 1134–1153 (1998)
Chan, C. W., Chetnani, B. & Mondragón, A. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip Rev. RNA http://dx.doi.org/10.1002/wrna.1175 (23 June 2013)
Edwards, T. E., Klein, D. J. & Ferré-D’Amaré, A. R. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 17, 273–279 (2007)
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006)
Rock, F. L. et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing eite. Science 316, 1759–1761 (2007)
Fukai, S. et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103, 793–803 (2000)
Dalluge, J. J., Hashizume, T., Sopchik, A. E., McCloskey, J. A. & Davis, D. R. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 24, 1073–1079 (1996)
Grigg, J. C. et al. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc. Natl Acad. Sci. USA 110, 7240–7245 (2013)
Krasilnikov, A. S., Yang, X., Pan, T. & Mondragón, A. Crystal structure of the specificity domain of ribonuclease P. Nature 421, 760–764 (2003)
Nikulin, A. et al. Structure of the L1 protuberance in the ribosome. Nature Struct. Biol. 10, 104–108 (2003)
Chang, A. T. & Nikonowicz, E. P. Solution nuclear magnetic resonance analyses of the anticodon arms of proteinogenic and nonproteinogenic tRNAGly. Biochemistry 51, 3662–3674 (2012)
Valle, M. et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nature Struct. Biol. 10, 899–906 (2003)
Dunkle, J. A. et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332, 981–984 (2011)
Schmeing, T. M. et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009)
Wang, J. & Nikonowicz, E. P. Solution structure of the K-turn and specifier loop domains from the Bacillus subtilis tyrS T-Box leader RNA. J. Mol. Biol. 408, 99–117 (2011)
Vitreschak, A. G., Mironov, A. A., Lyubetsky, V. A. & Gelfand, M. S. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14, 717–735 (2008)
Leontis, N. B. & Westhof, E. Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512 (2001)
Xiao, H., Edwards, T. E. & Ferré-D’Amaré, A. R. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem. Biol. 15, 1125–1137 (2008)
Baird, N. J. & Ferré-D'Amaré, A. R. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16, 598–609 (2010)
Kulshina, N., Edwards, T. E. & Ferré-D’Amaré, A. R. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA 16, 186–196 (2010)
Baird, N. J. & Ferré-D’Amaré, A. R. Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches. RNA 19, 167–176 (2013)
Schuck, P. Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000)
Keller, S. et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
Otwinowski, Z. & Minor, W. Processing of diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Pape, T. & Schneider, T. R. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 (2004)
Byrne, R. T., Konevega, A. L., Rodnina, M. V. & Antson, A. A. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 38, 4154–4162 (2010)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997)
Xiao, H., Murakami, H., Suga, H. & Ferré-D’Amaré, A. R. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature 454, 358–361 (2008)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Shi, H. & Moore, P. B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6, 1091–1105 (2000)
Correll, C. C., Freeborn, B., Moore, P. B. & Steitz, T. A. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell 91, 705–712 (1997)
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, 2002)
Lescoute, A., Leontis, N. B., Massire, C. & Westhof, E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 33, 2395–2409 (2005)
Acknowledgements
We thank the staff at beamlines 24-ID-C and 24-ID-E of APS and 5.0.1 and 5.0.2 of the ALS for crystallographic data collection support, G. Piszczek for ITC support, R. Levine and D.-Y. Lee for help with mass spectrometry, K. Perry and K. R. Rajashankar for assistance with initial crystallographic phasing, and N. Baird, T. Hamma, C. Jones, M. Lau, J. Posakony, A. Roll-Mecak, O. Uhlenbeck and K. Warner for discussions. This work is partly based on research conducted at the APS on the Northeastern Collaborative Access Team (NE-CAT) beamlines, and at the ALS on the Berkeley Center for Structural Biology beamlines, which are both supported by the National Institute for General Medical Sciences, National Institutes of Health (NIH). Use of ALS and APS was supported by the US Department of Energy. This work was supported in part by the intramural program of the National Heart, Lung, and Blood Institute, NIH.
Author information
Authors and Affiliations
Contributions
J.Z. and A.R.F.-D. designed experiments, J.Z. carried out all biochemistry and ITC experiments, J.Z. prepared co-crystals, J.Z. and A.R.F.-D. collected diffraction data, solved and refined the crystal structures, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Sequence of the B. subtilis glyQS T-box.
Unlike some other T-boxes, tRNAGly-responsive T-boxes (such as the B. subtilis glyQS T-box and the closely related O. iheyensis glyQ T-box) do not contain stems II and II A/B in the linker region. Conservation across known T-boxes is indicated in orange and yellow circles (from ref. 11). Base pairing between the specifier sequence in stem I and the antiterminator domain of the T-box and the anticodon and acceptor end, respectively, of tRNAGly is indicated. The functionally important K-turn8 is boxed. The sequences of tRNAGly of B. subtilis and O. iheyensis are identical.
Extended Data Figure 2 Representative isothermal calorimetric titrations.
a–f, Full-length tRNAGly (a–e) and an isolated tRNAGly ASL (f) were titrated into various truncated B. subtilis glyQS T-box stem I constructs (Methods). The specifier trinucleotide GGC is in bold. g, Superposition of the experimental data and fits for the experiments shown in Fig. 1a–c, colour-coded to correspond to schematic RNA construct depictions. h, ITC analysis of binding by the engineered tRNAGly construct used for co-crystallization. Results of all fits (Methods) are in Extended Data Table 1.
Extended Data Figure 3 Intermolecular interface of the stem-I–tRNA complex and phylogenetic conservation.
a, The residue 29–30 bulge allows the O. iheyensis stem I to bend and track the tRNA backbone. b, Structure21 of the equivalent region of the Geobacillus kaustophilus T-box, which folds as a C-loop52. c, Solvent-accessible surface coloured according to area buried from blue or white (no burial) to red (>75 Å2 per residue). d, Open-book view. e, Surfaces coloured by phylogenetic conservation of stem I residues (see Extended Data Fig. 1). Conservation of tRNA is not indicated. f, Open-book view. For comparison, RNase P binding to pre-tRNA buries6 2,959 Å2.
Extended Data Figure 4 T-box can accommodate bulky post-transcriptional modifications at tRNA position 37.
A86 of the T-box stem I is unstacked from the duplex formed by pairing of the tRNA anticodon and the T-box specifier by its participation in the sheared A•A pair at the base of the loop E motif. This results in a pocket (dotted line) that could accommodate the large, modified nucleobases (for example, the three-ring heterocycle wybutine in yeast tRNAPhe) often present at position 37 of tRNAs (Fig. 2a). Note the single hydrogen bond between tU32 and tA38.
Extended Data Figure 5 Structural organization of the interdigitated T-loop motifs of the T-box stem I.
a, Six base-paired layers form the stem I distal domain core (view as in Fig. 2b). Dashed black lines denote base–base hydrogen bonding. Dashed yellow lines denote Sr2+-ion–water (green and red spheres, respectively) interactions. Leontis–Westhof symbols30 depict non-canonical pairing. b, Orthogonal view. c, View from the tRNA elbow. d–i, Detail of layers 1–6. The position of each nucleotide within the pentaloop (1–5) or outside of it (negative and positive denote 5′ and 3′ directions, respectively) is in parentheses (Supplementary Discussion).
Extended Data Figure 6 Comparison of the interdigitated T-loops of RNase P, the L1 stalk and the T-box.
a, b, RNase P (a) and the L1 stalk (b) use equivalent surfaces formed by two purines—namely, nucleotide −2 of T-loop 1 and nucleotide 3 of T-loop 2 (asterisk)—to stack on the elbows of pre-tRNA and the E-site tRNA, respectively. c, By contrast, the T-box uses the opposite surface, using a triple (Fig. 2c) formed by nucleotide 3 of T-loop 1 (dagger symbol), nucleotide −2 from T-loop 2, and nucleotide +1 from T-loop 2 to stack on the tRNA elbow.
Extended Data Figure 7 Displacement of the tRNA elbow induced by T-box binding.
Although the anticodon stem of the free tRNAPhe (orange) superimposes well on that of the T-box-bound tRNAGly (T-loop and acceptor stem in violet; D-loop and ASL in pale green; Fig. 3b, c), the elbow (tG19–tC56) clashes with the stem I distal domain. T-box binding induces bending about the t26•t44 hinge (Fig. 3c) that displaces the tRNA elbow and acceptor stem away from the distal region of stem I by 11 Å (arrows), allowing the tRNA elbow to associate with the T-box. a, Side view. b, Top view.
Extended Data Figure 8 Representative electron density maps.
Portions of electron density maps corresponding to the interdigitated stem I T-loops and the tRNA elbow (oriented like Fig. 2b). a, Unbiased, 3.2-Å-resolution density-modified SAD electron density (1.0 s.d. above mean peak height) shown as grey mesh superimposed on the refined model. b, Electron density resulting from combination of SAD and molecular replacement (using a search model consisting of two copies each of tRNAPhe and a K-turn in complex with YbxF; Methods) phases followed by density modification (1.5 s.d.) c, Composite simulated anneal-omit 2|Fo| − |Fc| electron density calculated using the final model (2.0 s.d.).
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and additional references. (PDF 165 kb)
Rights and permissions
About this article
Cite this article
Zhang, J., Ferré-D’Amaré, A. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500, 363–366 (2013). https://doi.org/10.1038/nature12440
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12440
This article is cited by
-
Direct observation of tRNA-chaperoned folding of a dynamic mRNA ensemble
Nature Communications (2023)
-
Structural and dynamic mechanisms for coupled folding and tRNA recognition of a translational T-box riboswitch
Nature Communications (2023)
-
Streptococcus suis TrpX is part of a tryptophan uptake system, and its expression is regulated by a T-box regulatory element
Scientific Reports (2022)
-
Possible Ancestral Functions of the Genetic and RNA Operational Precodes and the Origin of the Genetic System
Origins of Life and Evolution of Biospheres (2021)
-
A SAM-I riboswitch with the ability to sense and respond to uncharged initiator tRNA
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.