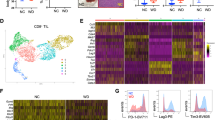
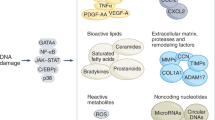
Abstract
Activated oncogenes and anticancer chemotherapy induce cellular senescence, a terminal growth arrest of viable cells characterized by S-phase entry-blocking histone 3 lysine 9 trimethylation (H3K9me3)1,2. Although therapy-induced senescence (TIS) improves long-term outcomes3, potentially harmful properties of senescent tumour cells make their quantitative elimination a therapeutic priority. Here we use the Eµ-myc transgenic mouse lymphoma model in which TIS depends on the H3K9 histone methyltransferase Suv39h1 to show the mechanism and therapeutic exploitation of senescence-related metabolic reprogramming in vitro and in vivo. After senescence-inducing chemotherapy, TIS-competent lymphomas but not TIS-incompetent Suv39h1– lymphomas show increased glucose utilization and much higher ATP production. We demonstrate that this is linked to massive proteotoxic stress, which is a consequence of the senescence-associated secretory phenotype (SASP) described previously4,5,6. SASP-producing TIS cells exhibited endoplasmic reticulum stress, an unfolded protein response (UPR), and increased ubiquitination, thereby targeting toxic proteins for autophagy in an acutely energy-consuming fashion. Accordingly, TIS lymphomas, unlike senescence models that lack a strong SASP response, were more sensitive to blocking glucose utilization or autophagy, which led to their selective elimination through caspase-12- and caspase-3-mediated endoplasmic-reticulum-related apoptosis. Consequently, pharmacological targeting of these metabolic demands on TIS induction in vivo prompted tumour regression and improved treatment outcomes further. These findings unveil the hypercatabolic nature of TIS that is therapeutically exploitable by synthetic lethal metabolic targeting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
20 August 2013
Source Data files for Figs 1–4 were added.
References
Narita, M. et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 (2003)
Braig, M. et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005)
Schmitt, C. A. et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109, 335–346 (2002)
Acosta, J. C. et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133, 1006–1018 (2008)
Coppe, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, e301 (2008)
Kuilman, T. et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 (2008)
Campisi, J. & d'Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nature Rev. Mol. Cell Biol. 8, 729–740 (2007)
Collado, M. & Serrano, M. Senescence in tumours: evidence from mice and humans. Nature Rev. Cancer 10, 51–57 (2010)
Kuilman, T., Michaloglou, C., Mooi, W. J. & Peeper, D. S. The essence of senescence. Genes Dev. 24, 2463–2479 (2010)
Nardella, C., Clohessy, J. G., Alimonti, A. & Pandolfi, P. P. Pro-senescence therapy for cancer treatment. Nature Rev. Cancer 11, 503–511 (2011)
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995)
Moskowitz, C. H. et al. Risk-adapted dose-dense immunochemotherapy determined by interim FDG-PET in advanced-stage diffuse large B-cell lymphoma. J. Clin. Oncol. 28, 1896–1903 (2010)
Jones, R. G. et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18, 283–293 (2005)
Warburg, O., Posener, K. & Negelein, E. Über den Stoffwechsel der Carcinomzelle. Biochem. Z. 152, 319–344 (1924)
Vander Heiden, M. G. et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329, 1492–1499 (2010)
Young, A. R. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009)
Chien, Y. et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011)
Jing, H. et al. Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev. 25, 2137–2146 (2011)
Kroemer, G., Marino, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010)
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/lymphoma cells3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007)
Nakagawa, T. et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403, 98–103 (2000)
Kaelin, W. G., Jr The concept of synthetic lethality in the context of anticancer therapy. Nature Rev. Cancer 5, 689–698 (2005)
Xue, W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007)
Kang, T. W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011)
Reimann, M. et al. Tumor stroma-derived TGF-β limits Myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17, 262–272 (2010)
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985)
Peters, A. H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001)
Schmitt, C. A. et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1, 289–298 (2002)
Schmitt, C. A., McCurrach, M. E., de Stanchina, E., Wallace-Brodeur, R. R. & Lowe, S. W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13, 2670–2677 (1999)
Shields, A. F. et al. Imaging proliferation in vivo with [F-18]FLT and positron emission tomography. Nature Med. 4, 1334–1336 (1998)
Marciniak, S. J. et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 (2004)
Reimann, M. et al. The Myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood 110, 2996–3004 (2007)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Walenta, S. et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 60, 916–921 (2000)
Liu, L. et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature 483, 608–612 (2012)
Kempa, S. et al. An automated GCxGC-TOF-MS protocol for batch-wise extraction and alignment of mass isotopomer matrixes from differential 13C-labelling experiments: a case study for photoautotrophic-mixotrophic grown Chlamydomonas reinhardtii cells. J. Basic Microbiol. 49, 82–91 (2009)
Giavalisco, P. et al. High-resolution direct infusion-based mass spectrometry in combination with whole 13C metabolome isotope labeling allows unambiguous assignment of chemical sum formulas. Anal. Chem. 80, 9417–9425 (2008)
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L. & Fernie, A. R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols 1, 387–396 (2006)
Cuadros-Inostroza, A. et al. TargetSearch—a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics 10, 428 (2009)
Lisec, J. et al. Corn hybrids display lower metabolite variability and complex metabolite inheritance patterns. Plant J. 68, 326–336 (2011)
Stacklies, W., Redestig, H., Scholz, M., Walther, D. & Selbig, J. pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–1167 (2007)
Bode, C. & Graler, M. H. Quantification of sphingosine-1-phosphate and related sphingolipids by liquid chromatography coupled to tandem mass spectrometry. Methods Mol. Biol. 874, 33–44 (2012)
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997)
Berns, K. et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431–437 (2004)
Reimer, T. A. et al. Reevaluation of the 22-1-1 antibody and its putative antigen, EBAG9/RCAS1, as a tumor marker. BMC Cancer 17, 47 (2005)
Castro, F. et al. High-throughput SNP-based authentication of human cell lines. Int. J. Cancer 132, 308–314 (2013)
Acknowledgements
We thank the late A. Harris, T. Jacks, T. Jenuwein, P. A. Khavari, N. Mizushima, D. Peeper, and M. Vander Heiden for mice, cells and materials; the flow cytometry facility at the Berlin-Brandenburg Center for Regenerative Therapies; N. Burbach, J. Dräger, A. Herrmann, K. Kirste, S. Maßwig, B. Teichmann and S. Spiesicke-Wegener for technical assistance; and members of the Schmitt laboratory for discussions and editorial advice. This work was supported by a Ph.D. fellowship to J.R.D. from the Boehringer Ingelheim Foundation, and grants from the Deutsche Forschungsgemeinschaft to W.M.-K. (MK576/15-1), to U.K. and A.K.B. (SFB 824), to U.K., B.D., S.L. and C.A.S. (SFB/TRR 54), and to C.A.S. from the Helmholtz Association (Helmholtz Alliance ‘Preclinical Comprehensive Cancer Center’; grant no. HA-305) and the Deutsche Krebshilfe (grant no. 108789). This interdisciplinary work was made possible by the structural framework of the inter-institutional cooperation between Charité and MDC (now represented by the Berlin Institute of Health (BIH)), the Berlin School of Integrative Oncology (BSIO) graduate program funded within the Excellence Initiative, and the German Cancer Consortium (GCC).
Author information
Authors and Affiliations
Contributions
J.R.D., S.L. and C.A.S. conceived the project, designed the experiments, and analysed the data, and W.M.-K., U.K., B.D., L.W. and St.K. provided critical input. Y.Y., G.B., C.Z., J.H.M.D., J.L., A.G., K.S., Su.K., S.W., M.G. and M.R. conducted experiments, M.M. compiled GSEA data, D.L. generated gene expression profiling data, M.H. analysed GEP data, B.P. carried out electron microscopy, and A.K.B. performed PET imaging. C.A.S., with editorial assistance from S.L., wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-16, Supplementary Table 1 and Supplementary References. (PDF 8227 kb)
Supplementary Data
This file contains source data for Supplementary Figure 1. (XLSX 49 kb)
Rights and permissions
About this article
Cite this article
Dörr, J., Yu, Y., Milanovic, M. et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425 (2013). https://doi.org/10.1038/nature12437
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12437
This article is cited by
-
Cholesterol biosynthetic pathway induces cellular senescence through ERRα
npj Aging (2024)
-
Blocking methionine catabolism induces senescence and confers vulnerability to GSK3 inhibition in liver cancer
Nature Cancer (2024)
-
MicroRNAs-associated with FOXO3 in cellular senescence and other stress responses
Biogerontology (2024)
-
Drug mechanism enrichment analysis improves prioritization of therapeutics for repurposing
BMC Bioinformatics (2023)
-
PDK4-dependent hypercatabolism and lactate production of senescent cells promotes cancer malignancy
Nature Metabolism (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.