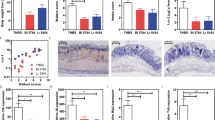
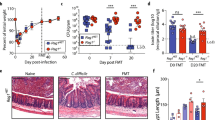
Abstract
Manipulation of the gut microbiota holds great promise for the treatment of inflammatory and allergic diseases1,2. Although numerous probiotic microorganisms have been identified3, there remains a compelling need to discover organisms that elicit more robust therapeutic responses, are compatible with the host, and can affect a specific arm of the host immune system in a well-controlled, physiological manner. Here we use a rational approach to isolate CD4+FOXP3+ regulatory T (Treg)-cell-inducing bacterial strains from the human indigenous microbiota. Starting with a healthy human faecal sample, a sequence of selection steps was applied to obtain mice colonized with human microbiota enriched in Treg-cell-inducing species. From these mice, we isolated and selected 17 strains of bacteria on the basis of their high potency in enhancing Treg cell abundance and inducing important anti-inflammatory molecules—including interleukin-10 (IL-) and inducible T-cell co-stimulator (ICOS)—in Treg cells upon inoculation into germ-free mice. Genome sequencing revealed that the 17 strains fall within clusters IV, XIVa and XVIII of Clostridia, which lack prominent toxins and virulence factors. The 17 strains act as a community to provide bacterial antigens and a TGF-β-rich environment to help expansion and differentiation of Treg cells. Oral administration of the combination of 17 strains to adult mice attenuated disease in models of colitis and allergic diarrhoea. Use of the isolated strains may allow for tailored therapeutic manipulation of human immune disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nature Rev. Immunol. 9, 313–323 (2009)
Honda, K. & Littman, D. R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 30, 759–795 (2012)
O’Toole, P. W. & Cooney, J. C. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip. Perspect. Infect. Dis. 2008, 175285 (2008)
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011)
Geuking, M. B. et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806 (2011)
Russell, S. L. et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447 (2012)
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010)
Chung, H. et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593 (2012)
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008)
Thornton, A. M. et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184, 3433–3441 (2010)
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008)
Wing, K. et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008)
Zheng, Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature 458, 351–356 (2009)
Maslowski, K. M. & Mackay, C. R. Diet, gut microbiota and immune responses. Nature Immunol. 12, 5–9 (2011)
Lathrop, S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011)
Cebula, A. et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 497, 258–262 (2013)
Collins, M. D. et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44, 812–826 (1994)
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010)
Kweon, M. N., Yamamoto, M., Kajiki, M., Takahashi, I. & Kiyono, H. Systemically derived large intestinal CD4+ Th2 cells play a central role in STAT6-mediated allergic diarrhea. J. Clin. Invest. 106, 199–206 (2000)
Strober, W., Fuss, I. J. & Blumberg, R. S. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 20, 495–549 (2002)
Lawley, T. D. et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995 (2012)
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007)
Manichanh, C. et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55, 205–211 (2006)
Candela, M. et al. Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 12, 95 (2012)
Morita, H. et al. An improved isolation method for metagenomic analysis of the microbial flora of the human intestine. Microbes Environ. 22, 214–222 (2007)
Kim, S. W. et al. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 20, 241–253 (2013)
Noguchi, H., Taniguchi, T. & Itoh, T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 15, 387–396 (2008)
Jolley, K. A. et al. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158, 1005–1015 (2012)
Acknowledgements
This work was supported by JSPS NEXT program, Grant in Aid for Scientific Research on Innovative Areas ‘Genome Science’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No.221S0002), the global COE project of ‘Genome Information Big Bang’ and the Waksman Foundation of Japan Inc. We thank M. Suyama, K. Furuya, C. Yoshino, H. Inaba, E. Iioka, Y. Takayama, M. Kiuchi, Y. Hattori, N. Fukuda and A. Nakano for technical assistance, and P. D. Burrows for review of the manuscript.
Author information
Authors and Affiliations
Contributions
K.Ho. planned experiments, analysed data and wrote the paper together with B.O. and M.H.; K.A. and T.Tano. performed immunological analyses and bacterial cultures together with Y.N., S.N. and H.M.; W.S., K.O., S.K. and M.H. performed bacterial sequence analyses; K.M. and S.U. provided essential materials; H.N., T.S. and S.S. supervised the Treg cell suppression assay; S.F., K.Ha., H.O., T.Tani., J.V.F. and P.W. were involved in data discussions.
Corresponding authors
Ethics declarations
Competing interests
B.O. is an employee of PureTech Ventures.
Additional information
All genome sequence data are deposited in DDBJ BioProject ID PRJDB521-543.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-17. (PDF 1797 kb)
Supplementary Table 1
This file contains meta 16S rRNA gene analysis for the series of gnotobiotic mice. The numbers of detected reads, the closest species, and % similarities with the closest species for each OTU in each exGF mouse are shown. (XLSX 32 kb)
Supplementary Table 2
This file contains putative toxins and virulence factors found in 17 strains. BLASTP search of gene products predicted from genomes was performed using virulence factor databases (VFDB and MvirDB) with the e-value cut off of 1.0e-10, the identity >30% and the length coverage >60%. Note that several strains possess genes encoding putative hyaluronidase, sialidase, fibronectin-binding proteins, and flagella-related proteins but with low similarity to genes of pathogenic Clostridia species, and most of these genes are also encoded by other commensal Clostridia species. (XLSX 72 kb)
Rights and permissions
About this article
Cite this article
Atarashi, K., Tanoue, T., Oshima, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013). https://doi.org/10.1038/nature12331
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12331
This article is cited by
-
Analysis of differences in intestinal flora associated with different BMI status in colorectal cancer patients
Journal of Translational Medicine (2024)
-
Lung microbiome: new insights into the pathogenesis of respiratory diseases
Signal Transduction and Targeted Therapy (2024)
-
Enhancing tumor-specific recognition of programmable synthetic bacterial consortium for precision therapy of colorectal cancer
npj Biofilms and Microbiomes (2024)
-
Evaluation of regulatory T-cells in cancer immunotherapy: therapeutic relevance of immune checkpoint inhibition
Medical Oncology (2024)
-
The Potential of Clostridium butyricum to Preserve Gut Health, and to Mitigate Non-AIDS Comorbidities in People Living with HIV
Probiotics and Antimicrobial Proteins (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.