Abstract
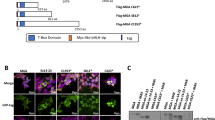
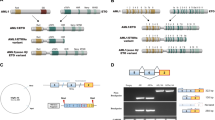
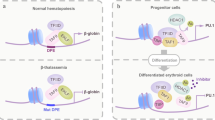
Transcription factors are frequently altered in leukaemia through chromosomal translocation, mutation or aberrant expression1. AML1–ETO, a fusion protein generated by the t(8;21) translocation in acute myeloid leukaemia, is a transcription factor implicated in both gene repression and activation2. AML1–ETO oligomerization, mediated by the NHR2 domain, is critical for leukaemogenesis3,4,5,6, making it important to identify co-regulatory factors that ‘read’ the NHR2 oligomerization and contribute to leukaemogenesis4. Here we show that, in human leukaemic cells, AML1–ETO resides in and functions through a stable AML1–ETO-containing transcription factor complex (AETFC) that contains several haematopoietic transcription (co)factors. These AETFC components stabilize the complex through multivalent interactions, provide multiple DNA-binding domains for diverse target genes, co-localize genome wide, cooperatively regulate gene expression, and contribute to leukaemogenesis. Within the AETFC complex, AML1–ETO oligomerization is required for a specific interaction between the oligomerized NHR2 domain and a novel NHR2-binding (N2B) motif in E proteins. Crystallographic analysis of the NHR2–N2B complex reveals a unique interaction pattern in which an N2B peptide makes direct contact with side chains of two NHR2 domains as a dimer, providing a novel model of how dimeric/oligomeric transcription factors create a new protein-binding interface through dimerization/oligomerization. Intriguingly, disruption of this interaction by point mutations abrogates AML1–ETO-induced haematopoietic stem/progenitor cell self-renewal and leukaemogenesis. These results reveal new mechanisms of action of AML1–ETO, and provide a potential therapeutic target in t(8;21)-positive acute myeloid leukaemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Look, A. T. Oncogenic transcription factors in the human acute leukemias. Science 278, 1059–1064 (1997)
Peterson, L. F. & Zhang, D. E. The 8;21 translocation in leukemogenesis. Oncogene 23, 4255–4262 (2004)
Minucci, S. et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol. Cell 5, 811–820 (2000)
Liu, Y. et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell 9, 249–260 (2006)
Kwok, C., Zeisig, B. B., Qiu, J., Dong, S. & So, C. W. Transforming activity of AML1-ETO is independent of CBFβ and ETO interaction but requires formation of homo-oligomeric complexes. Proc. Natl Acad. Sci. USA 106, 2853–2858 (2009)
Yan, M., Ahn, E. Y., Hiebert, S. W. & Zhang, D. E. RUNX1/AML1 DNA-binding domain and ETO/MTG8 NHR2-dimerization domain are critical to AML1-ETO9a leukemogenesis. Blood 113, 883–886 (2009)
Okumura, A. J., Peterson, L. F., Okumura, F., Boyapati, A. & Zhang, D. E. t(8;21)(q22;q22) fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood 112, 1392–1401 (2008)
Gelmetti, V. et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 18, 7185–7191 (1998)
Lutterbach, B. et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18, 7176–7184 (1998)
Wang, J., Hoshino, T., Redner, R. L., Kajigaya, S. & Liu, J. M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl Acad. Sci. USA 95, 10860–10865 (1998)
Westendorf, J. J. et al. The t(8;21) fusion product, AML-1–ETO, associates with C/EBP-α, inhibits C/EBP-α-dependent transcription, and blocks granulocytic differentiation. Mol. Cell. Biol. 18, 322–333 (1998)
Mao, S., Frank, R. C., Zhang, J., Miyazaki, Y. & Nimer, S. D. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Mol. Cell. Biol. 19, 3635–3644 (1999)
Elagib, K. E. et al. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101, 4333–4341 (2003)
Zhang, J., Kalkum, M., Yamamura, S., Chait, B. T. & Roeder, R. G. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305, 1286–1289 (2004)
Gardini, A. et al. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 4, e1000275 (2008)
Guo, C., Hu, Q., Yan, C. & Zhang, J. Multivalent binding of the ETO corepressor to E proteins facilitates dual repression controls targeting chromatin and the basal transcription machinery. Mol. Cell. Biol. 29, 2644–2657 (2009)
Martens, J. H. et al. ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood 120, 4038–4048 (2012)
Wang, L. et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 333, 765–769 (2011)
Shia, W. J. et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 119, 4953–4962 (2012)
Miyoshi, H. et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 12, 2715–2721 (1993)
Wadman, I. A. et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16, 3145–3157 (1997)
Follows, G. A. et al. Epigenetic consequences of AML1-ETO action at the human c-FMS locus. EMBO J. 22, 2798–2809 (2003)
Huang, G. et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nature Genet. 40, 51–60 (2008)
Heidenreich, O. et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood 101, 3157–3163 (2003)
Yan, M. et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nature Med. 12, 945–949 (2006)
Sanchez, M. et al. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development 126, 3891–3904 (1999)
Park, S. et al. Structure of the AML1-ETO eTAFH domain–HEB peptide complex and its contribution to AML1-ETO activity. Blood 113, 3558–3567 (2009)
Mulloy, J. C. et al. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 99, 15–23 (2002)
Amoutzias, G. D., Robertson, D. L., Van de, P. Y. & Oliver, S. G. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 33, 220–229 (2008)
Funnell, A. P. & Crossley, M. Homo- and heterodimerization in transcriptional regulation. Adv. Exp. Med. Biol. 747, 105–121 (2012)
Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983)
Giannopoulou, E. G. & Elemento, O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics 12, 277 (2011)
Sharov, A. A. & Ko, M. S. Exhaustive search for over-represented DNA sequence motifs with CisFinder. DNA Res. 16, 261–273 (2009)
Ji, H. et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nature Biotechnol. 26, 1293–1300 (2008)
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 (2008)
Jacobs, S. A., Fischle, W. & Khorasanizadeh, S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 376, 131–148 (2004)
Zuber, J. et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 23, 877–889 (2009)
Acknowledgements
We thank N. A. Speck and J. H. Bushweller for providing the AML1–ETO m7 plasmid, and R. Baer for providing anti-SCL antibodies. This work was supported by National Institutes of Health (NIH) grants CA163086 (R.G.R.), CA129325 (R.G.R.), CA113872 (R.G.R.) and CA166835 (S.D.N.), Starr Cancer Consortium grant I5-A554 (R.G.R., D.J.P. and S.D.N.), Leukemia and Lymphoma Society (LLS) SCOR grants 7013-02 (R.G.R. and S.D.N.) and 7132-08 (R.G.R., A.M. and D.J.P.), and Rockefeller University Center for Clinical and Translational Science Pilot Project grant UL1RR024143 from NIH (X.-J.S.). X.-J.S. was a Starr Cancer Consortium Visiting Fellow. L.W. was an Empire State Stem Cell Scholar and an LLS Fellow. Y.J. was an American Society of Haematology Scholar. W.-Y.C. was an LLS Fellow. D.J.P. was supported by funds from the Abby Rockefeller Mauze Trust and the Maloris Foundation.
Author information
Authors and Affiliations
Contributions
X.-J.S. and R.G.R. conceived the project. R.G.R. supervised the biochemical studies. S.D.N. supervised the leukaemia pathological studies. D.J.P. supervised the structural studies. A.M. supervised the genomic studies. X.-J.S., Z.W., L.W., Y.J., N.K., T.D.S., W.-Y.C., Z.T., T.N., O.E. and W.F. performed the experiments and analysed the data. X.J.S. and R.G.R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-15, Supplementary Tables 1-3, a Supplementary Discussion and additional references. (PDF 2895 kb)
Rights and permissions
About this article
Cite this article
Sun, XJ., Wang, Z., Wang, L. et al. A stable transcription factor complex nucleated by oligomeric AML1–ETO controls leukaemogenesis. Nature 500, 93–97 (2013). https://doi.org/10.1038/nature12287
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12287
This article is cited by
-
A novel AML1-ETO/FTO positive feedback loop promotes leukemogenesis and Ara-C resistance via stabilizing IGFBP2 in t(8;21) acute myeloid leukemia
Experimental Hematology & Oncology (2024)
-
Combination of eriocalyxin B and homoharringtonine exerts synergistic anti-tumor effects against t(8;21) AML
Acta Pharmacologica Sinica (2024)
-
LMO2 promotes the development of AML through interaction with transcription co-regulator LDB1
Cell Death & Disease (2023)
-
Myeloid neoplasms and clonal hematopoiesis from the RUNX1 perspective
Leukemia (2022)
-
Biophysical and pharmacokinetic characterization of a small-molecule inhibitor of RUNX1/ETO tetramerization with anti-leukemic effects
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.