Abstract
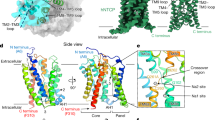
The hepatitis C virus (HCV) has developed a small membrane protein, p7, which remarkably can self-assemble into a large channel complex that selectively conducts cations1,2,3,4. We wanted to examine the structural solution that the viroporin adopts in order to achieve selective cation conduction, because p7 has no homology with any of the known prokaryotic or eukaryotic channel proteins. The activity of p7 can be inhibited by amantadine and rimantadine2,5, which are potent blockers of the influenza M2 channel6 and licensed drugs against influenza infections7. The adamantane derivatives have been used in HCV clinical trials8, but large variation in drug efficacy among the various HCV genotypes has been difficult to explain without detailed molecular structures. Here we determine the structures of this HCV viroporin as well as its drug-binding site using the latest nuclear magnetic resonance (NMR) technologies. The structure exhibits an unusual mode of hexameric assembly, where the individual p7 monomers, i, not only interact with their immediate neighbours, but also reach farther to associate with the i+2 and i+3 monomers, forming a sophisticated, funnel-like architecture. The structure also points to a mechanism of cation selection: an asparagine/histidine ring that constricts the narrow end of the funnel serves as a broad cation selectivity filter, whereas an arginine/lysine ring that defines the wide end of the funnel may selectively allow cation diffusion into the channel. Our functional investigation using whole-cell channel recording shows that these residues are critical for channel activity. NMR measurements of the channel–drug complex revealed six equivalent hydrophobic pockets between the peripheral and pore-forming helices to which amantadine or rimantadine binds, and compound binding specifically to this position may allosterically inhibit cation conduction by preventing the channel from opening. Our data provide a molecular explanation for p7-mediated cation conductance and its inhibition by adamantane derivatives.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Moradpour, D., Penin, F. & Rice, C. M. Replication of hepatitis C virus. Nature Rev. Microbiol. 5, 453–463 (2007)
Griffin, S. D. et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535, 34–38 (2003)
Pavlovic, D. et al. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl Acad. Sci. USA 100, 6104–6108 (2003)
Luik, P. et al. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl Acad. Sci. USA 106, 12712–12716 (2009)
Griffin, S. et al. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology 48, 1779–1790 (2008)
Wang, C., Takeuchi, K., Pinto, L. H. & Lamb, R. A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 67, 5585–5594 (1993)
Davies, W. L. et al. Antiviral activity of 1-sdamantanamine (amantadine). Science 144, 862–863 (1964)
Mihm, U. et al. Amino acid variations in hepatitis C virus p7 and sensitivity to antiviral combination therapy with amantadine in chronic hepatitis C. Antivir. Ther. 11, 507–519 (2006)
Fischer, W. B. & Sansom, M. S. Viral ion channels: structure and function. Biochim. Biophys. Acta 1561, 27–45 (2002)
Nieva, J. L., Madan, V. & Carrasco, L. Viroporins: structure and biological functions. Nature Rev. Microbiol. 10, 563–574 (2012)
Steinmann, E. et al. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology 46, 330–338 (2007)
Montserret, R. et al. NMR structure and ion channel activity of the p7 protein from hepatitis C virus. J. Biol. Chem. 285, 31446–31461 (2010)
Premkumar, A., Wilson, L., Ewart, G. D. & Gage, P. W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 557, 99–103 (2004)
Wozniak, A. L. et al. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6, e1001087 (2010)
Sakai, A. et al. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl Acad. Sci. USA 100, 11646–11651 (2003)
Jones, C. T., Murray, C. L., Eastman, D. K., Tassello, J. & Rice, C. M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81, 8374–8383 (2007)
Steinmann, E. et al. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3, e103 (2007)
Popescu, C. I. et al. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7, e1001278 (2011)
Vieyres, G. et al. Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells. J. Virol. 87, 1664–1678 (2013)
Cook, G. A. & Opella, S. J. Secondary structure, dynamics, and architecture of the p7 membrane protein from hepatitis C virus by NMR spectroscopy. Biochim. Biophys. Acta 1808, 1448–1453 (2011)
Stouffer, A. L. et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 451, 596–599 (2008)
Cady, S. D. et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 463, 689–692 (2010)
Pielak, R. M., Oxenoid, K. & Chou, J. J. Structural investigation of rimantadine inhibition of the AM2–BM2 chimera channel of influenza viruses. Structure 19, 1655–1663 (2011)
Oxenoid, K. & Chou, J. J. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc. Natl Acad. Sci. USA 102, 10870–10875 (2005)
Schnell, J. R. & Chou, J. J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451, 591–595 (2008)
Van Horn, W. D. et al. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science 324, 1726–1729 (2009)
Hou, X., Pedi, L., Diver, M. M. & Long, S. B. Crystal structure of the calcium release-activated calcium channel Orai. Science 338, 1308–1313 (2012)
StGelais, C. et al. Determinants of hepatitis C virus p7 ion channel function and drug sensitivity identified in vitro. J. Virol. 83, 7970–7981 (2009)
Foster, T. L. et al. Resistance mutations define specific antiviral effects for inhibitors of the hepatitis C virus p7 ion channel. Hepatology 54, 79–90 (2011)
Cuello, L. G., Jogini, V., Cortes, D. M. & Perozo, E. Structural mechanism of C-type inactivation in K+ channels. Nature 466, 203–208 (2010)
Pervushin, K., Riek, R., Wider, G. & Wuthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA 94, 12366–12371 (1997)
Kay, L. E., Torchia, D. A. & Bax, A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28, 8972–8979 (1989)
Szyperski, T., Neri, D., Leiting, B., Otting, G. & Wuthrich, K. Support of 1H NMR assignments in proteins by biosynthetically directed fractional 13C-labeling. J. Biomol. NMR 2, 323–334 (1992)
Chou, J. J., Gaemers, S., Howder, B., Louis, J. M. & Bax, A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR 21, 377–382 (2001)
Sass, H. J., Musco, G., Stahl, S. J., Wingfield, P. T. & Grzesiek, S. Solution NMR of proteins within polyacrylamide gels: Diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J. Biomol. NMR 18, 303–309 (2000)
Tycko, R., Blanco, F. J. & Ishii, Y. Alignment of biopolymers in strained gels: A new way to create detectable dipole-dipole couplings in high-resolution biomolecular NMR. J. Am. Chem. Soc. 122, 9340–9341 (2000)
Weigelt, J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 120, 10778–10779 (1998)
Wu, J., Fan, J. S., Pascal, S. M. & Yang, D. General method for suppression of diagonal peaks in heteronuclear-edited NOESY spectroscopy. J. Am. Chem. Soc. 126, 15018–15019 (2004)
Schwieters, C. D., Kuszewski, J., Tjandra, N. & Clore, G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 66–74 (2002)
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. W. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993)
Plugge, B. et al. A potassium channel protein encoded by chlorella virus PBCV-1. Science 287, 1641–1644 (2000)
Acknowledgements
We thank R. Sounier for helping with making specific methyl-labelled protein, S. Brueschweiler for helping with ITC measurements, G. Bellot, J. Min and W. Shih for providing DNA nanotube liquid crystal, and K. Oxenoid for discussion. This work was supported by the National Key Project of 973 (2013CB530504) and National Science and Technology Major Project (2012ZX10002-007-003) (to B.S.) and NIH grant GM094608 (to J.J.C.).
Author information
Authors and Affiliations
Contributions
B.O. and J.J.C. conceived the study; B.O. prepared samples; M.J.B. performed EM analysis; J.D. and B.O. performed NMR titration; B.O. and J.J.C. collected and analysed NMR data and determined the structure; S.X., X.Z., W.Y. and B.S. designed and performed functional experiments; J.J.C. wrote the paper and all authors contributed to the editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This fie contains Supplementary Methods, Supplementary Table 1, Supplementary Figures 1-11 and additional references. (PDF 11093 kb)
Rights and permissions
About this article
Cite this article
OuYang, B., Xie, S., Berardi, M. et al. Unusual architecture of the p7 channel from hepatitis C virus. Nature 498, 521–525 (2013). https://doi.org/10.1038/nature12283
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12283
This article is cited by
-
Experimental and computational studies of tautomerism pyridine carbonyl thiosemicarbazide derivatives
Structural Chemistry (2023)
-
Amantadine inhibits known and novel ion channels encoded by SARS-CoV-2 in vitro
Communications Biology (2021)
-
Identification of Human Secretome and Membrane Proteome-Based Cancer Biomarkers Utilizing Bioinformatics
The Journal of Membrane Biology (2020)
-
Progresses in Predicting Post-translational Modification
International Journal of Peptide Research and Therapeutics (2020)
-
Hepatitis C virus sequence divergence preserves p7 viroporin structural and dynamic features
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.